

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_0612000 | SP | 0.018848 | 0.980647 | 0.000505 | CS pos: 17-18. CVC-KK. Pr: 0.9104 |
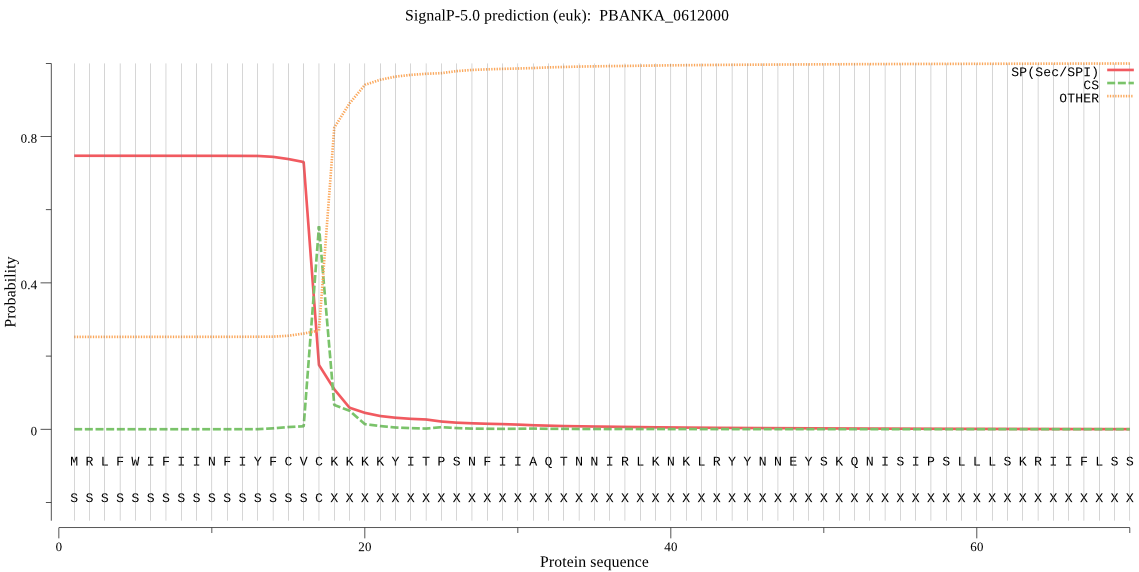
MRLFWIFIINFIYFCVCKKKKYITPSNFIIAQTNNIRLKNKLRYYNNEYSKQNISIPSLL LSKRIIFLSSPIYPHISEQIISQLLYLEYESKRKPIHLYINSTGDLENNKIVNLNGITDV ISIIDVINYISSDVYTYCLGKAYGIACILASSGKKGYRFSLKNSSFCLKQSYSIIPFNQA TNIEIQNKEIMNTKKKVIDIIAKNTEKDNNIISEILDRDRYFNANEASDFNLIDHILEKQ