

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
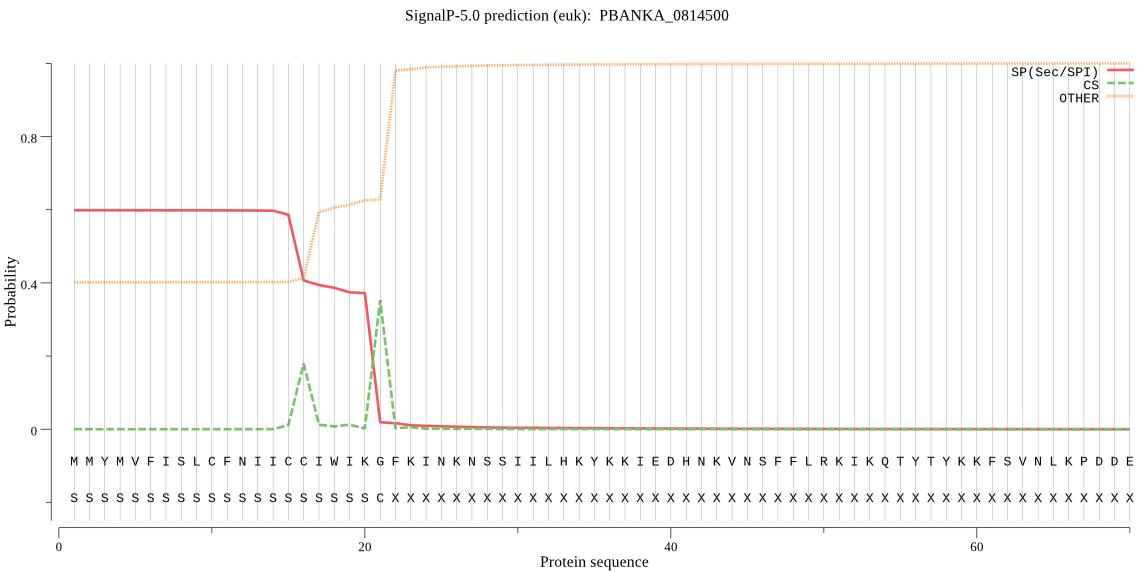
| PBANKA_0814500 | OTHER | 0.658885 | 0.337986 | 0.003130 |
MMYMVFISLCFNIICCIWIKGFKINKNSSIILHKYKKIEDHNKVNSFFLRKIKQTYTYKK FSVNLKPDDESVNNIKERKNIIKENIFNFYKKNIYLFSKILLYMKNGATFLTEKCTSSII PQLLISISFFFLHFYLLSKNFIILLPYQLIPNYSNILMGLDFNSTFFIFSSLFFLKNFIS NFQSFKTRLASNKFTIPLKENKKKNILTISVLFLTSYILSGYVSIYAEQILNYSKSVNFR ISDTTIKSLQILIGHFVWVGCSIKIFSKFLYPHFKNNISNLNFRYKDSWCFKVIYGYVCS HYIFNIIDLLNNTIINYFNNDDEIYIENSIDDIINEKKFLSTFLCVISPCFSAPFFEEFI YRFFVLKSLCLFMNIHYAVTFSSLLFAIHHLNIFNVIPLFFLSFFWSYIYIYTDNILITM IIHSFWNIYVFLSSLYN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PBANKA_0814500 | 191 S | TRLASNKFT | 0.994 | unsp |