

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_0906000 | OTHER | 0.972132 | 0.000245 | 0.027623 |
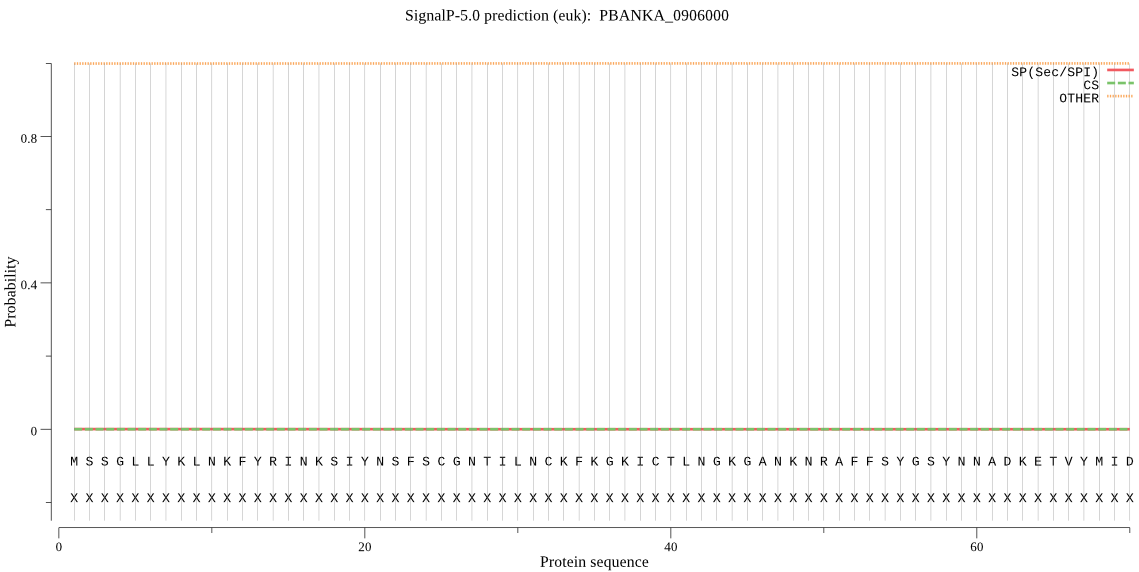
MSSGLLYKLNKFYRINKSIYNSFSCGNTILNCKFKGKICTLNGKGANKNRAFFSYGSYNN ADKETVYMIDGISFTMRNNSSIYKEETNDIPILLLHGCYGSKKNFRNFNKMLKSNKIISL DLPNHGESKHTDNMKYNNIEEDIKNVLSKLNIKSCCLVGFSLGGKVSMYTALKNPGLFSH LIIMDILPYNYNSNNIEIALPYSIKKMSKTLYNIKINKNPKNKFEFLTYLKEELPDIPDS FSQFICMSLKENEQKNKLTWNINVETIYNEIHNITNFPLNYEKYKYMNPCSFVIAKKSDL AYTIPDYDQIIKNFFPFSKNFILENSSHAVYVDAPYECAQIINQTITL