

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
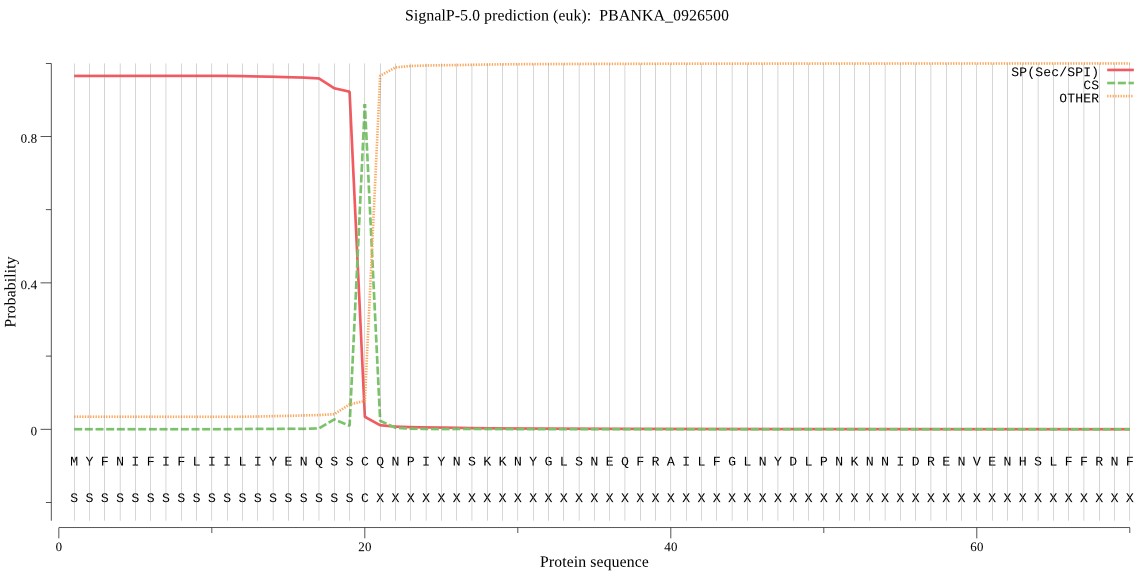
| PBANKA_0926500 | SP | 0.018491 | 0.981447 | 0.000062 | CS pos: 20-21. SSC-QN. Pr: 0.7362 |
MYFNIFIFLIILIYENQSSCQNPIYNSKKNYGLSNEQFRAILFGLNYDLPNKNNIDRENV ENHSLFFRNFVDDDIQINKQNEKTQSQIEIMNNEKIKSSDTIHDNKNVNNYSITSGNEKT NDGLLQKTLRKNKLSEKDNSYNNTSTNINKNENSVFNEPTNANDYIRTKEDSNDFKPETL QKNINVKNNKGNPNLLKDKKTNINENKTQGNINQNTRINKSVNDMTNELDNHIENFVKEG NEMNNSNVQEKHQGKDLLHNNGNNKDTQMDHNDEPQILSNGKKKKNTKYISMNPRAIKHK EENVNYSPKKIISGLPNSGNGVLKEIQPVKYNENINKKLNDASKGNENLQKTTNTDENIF SENITLKNITNDTNYYKYFKLKENGFRGLGIINKYSSKGCFSISVDCGRYNDLDDIPGIS NLLQRAIFYKSKKRDTTLLSELGVNSSKYNSHINESFTNFYASGNSEDIYYLLNLFVQNL FYPIFNEESIENEVNEISNKYISMENNPETCIKITSQYLTHFKYSNFFIYGNYITLCENI LKKKIDIKKKLYEFHRKCYQPKNMSISILLGKRSNSSDHYNINDIMNMVVQFFGKIKNYN YEENDIKKQNNMKLEENLSNRQVNNYNNRIFTYNDAQINLNEEINNKNDMSIPFINKLKY ALDLNQKSKYIEILKKEGWENQIFLYWSSKINIYIYKKIEEFKVMRFFRELFSNFRKDGL YYKISVENEYAYDFQIINICNKYYLNYGILIKLTEKGKSNIAHLIYIFQTFINEISKLFD HDSLNKGVNKYILDYYRETTLTTNINFRKDGINICLNDLINYSNTLLIYLENPLEFLTNN NLVENVNKNGFRNEIKITSLIGSLIRNENMHIINVVDKFTITNQIRIPNTSIEYSIGDNP YIIDEGKITNNINFTFPEFKICPFSNFKKNAILNENSFFCVSYNSKENFNYSKSNKQTFV SDDNEYVKSSILYNIPCLIKSSYGYNIFFKKGLTETSRVNADFIFFFPSKNFTFYEAIFT RIHIIILKKKIKQMLSDYTNCSVNVNIKDNVESYILHVDSNSYYFGDMLNKIEDLLSIKE VPTSDEFNYAYDELNLYVKRKDNVVVGDSLNIIHSLFNKYIPTNKEIYDILNAYFFYPLY NSYIKYINNFFHKNYINIFIYGNLSIPNEINIKNETNYGINTEYNSTNSVINNNNGVNYN TYSYINNTYTMYNKHALGKGDNMLEDKEENIHEEYKENSIEILQNGIGIKYIIDLCESFI RNVTNNVIRKSESTYYATKLINNEDIEIDIQIPDKNVGNSSITVSYIIESETILSDMLIN IIVDLISSDFIKFAKIKYNDGYTVDVKTLSTKYGFGGIIFVIQSFDNDVEKLEEDICGFV KHLTFQLMNIDIYDLVKKMQYMKKQYILNNTIFTFNQEYSTILDEIINGNECFDKKYKIV KIFDELINCPKIILNKANYILQNAKKLIFKEYKTSNAPNNVNEQMNYIYSNKRCNYSHNK NDILSNIELSNTVNFTKATTLNNGVPNYNVFRMNNMHKNWNYIINVSNFLEIKRKGFVQY VIDYFKNPYKLSLNHNNYLDYKSCDDEMHKDNFHVFHNFINDINEIREYFLAKFSNDQEN KEKCSINYEEIKKHCYEVNARMYG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0926500 | 503 S | NKYISMENN | 0.996 | unsp | PBANKA_0926500 | 503 S | NKYISMENN | 0.996 | unsp | PBANKA_0926500 | 503 S | NKYISMENN | 0.996 | unsp | PBANKA_0926500 | 895 S | SIEYSIGDN | 0.994 | unsp | PBANKA_0926500 | 1077 S | EDLLSIKEV | 0.994 | unsp | PBANKA_0926500 | 1084 S | EVPTSDEFN | 0.993 | unsp | PBANKA_0926500 | 135 S | KNKLSEKDN | 0.998 | unsp | PBANKA_0926500 | 431 S | IFYKSKKRD | 0.991 | unsp |