

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
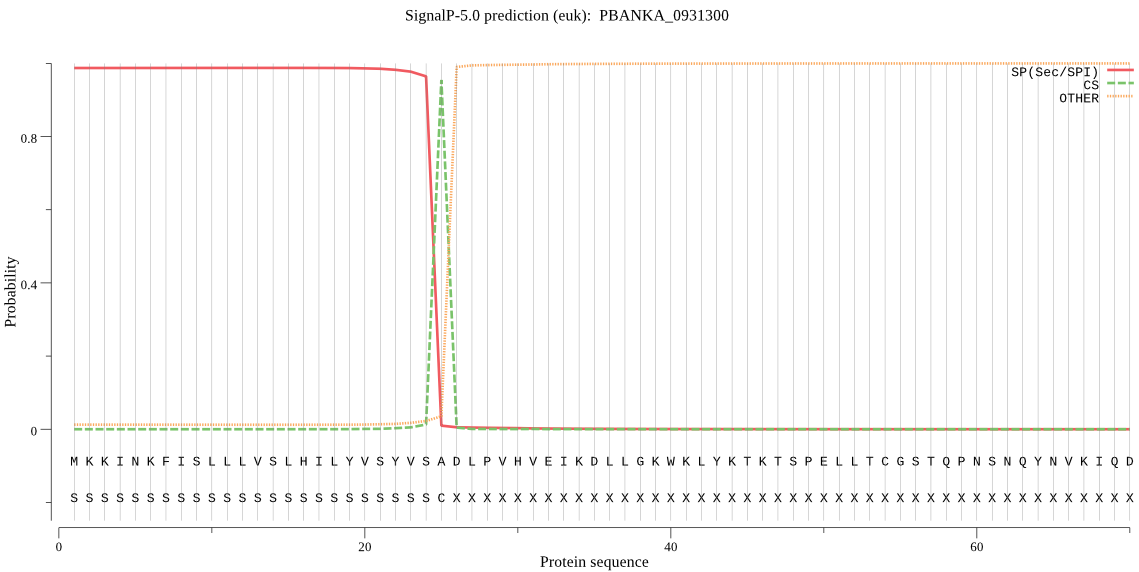
| PBANKA_0931300 | SP | 0.001621 | 0.998369 | 0.000010 | CS pos: 25-26. VSA-DL. Pr: 0.9689 |
MKKINKFISLLLVSLHILYVSYVSADLPVHVEIKDLLGKWKLYKTKTSPELLTCGSTQPN SNQYNVKIQDYKKYLTDNHYPFDSELNVILSSDFVKYGDVHDITDNEHRENWNVLAVYDE QKRKKIGTWTTIFDQGFEIRIGNETYTAFMHYEPTGKCAEPSDEDITDSNGETICYSTSY DKTRFGWIDIMNKNNEQLHGCFYAEKYDTLHVSNNYKNILNRFSNGKTEPVITEDKICTS NQITDFDSYEPKTYTKSNKVKLNKNSEMYWHKMKHDGKKKPLPEYMLKAQNQKYACPCNS NESIDNERSNEDPDSPVSPNMIELGNSNVDTNELDLNAYEEIKKSKHTELELNEMPKNFT WGDPFNNNVREYEVIDQLTCGSCYIASQMYVIKRRIEIGLTKLLETKYANDFDDALSLQT VLSCSFYDQGCHGGYPFLVSKMAKLHGIPLNSEFPYTAKQSTCPYPVNKGIPLSMMEVDL INKTESIPKYIKPSFRETNTSSQIENNTKEEINNVIDSGDPNRWYIKEYNYVGGCYGCNQ CDGEKIIMNEIYRNGPVVGSIEVTPNFYNYVDGVYYDKGFPHAKKCTVDVNKDNGYVYNI TGWEKVNHAIVILGWGEEIIDGKLYKYWICRNSWGNYWGKEGYFKMIRGVNYVAIENHAI YIDPDFTRGAGKVLLEKMKKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0931300 | 162 S | CAEPSDEDI | 0.995 | unsp | PBANKA_0931300 | 309 S | DNERSNEDP | 0.997 | unsp |