

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_0931800 | OTHER | 0.941273 | 0.045584 | 0.013143 |
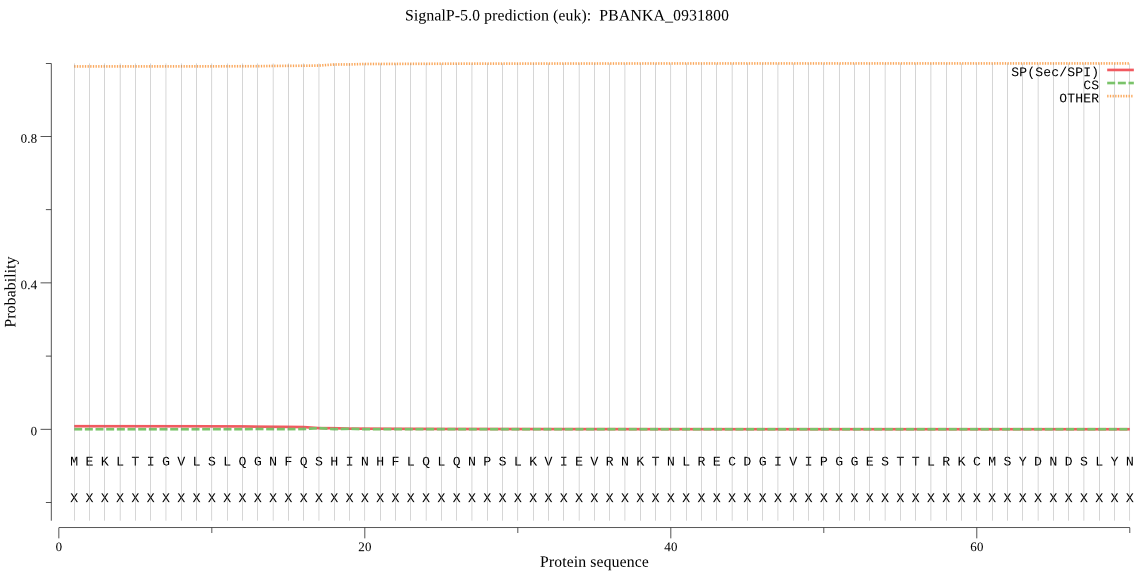
MEKLTIGVLSLQGNFQSHINHFLQLQNPSLKVIEVRNKTNLRECDGIVIPGGESTTLRKC MSYDNDSLYNALKNYIHVKKKPVWGTCAGCILLSEKVEKNKDDNIENEYGNDFSLGGLDI EITRNYYGSQNDSFICSLDIKSQDPIFKKNIRAPCIRAPFIKKISSDKVVTIATFSHESF GKNIIGAVEQDNCMGTIFHPELMPYTCFHDYFLEKVKKHIKDSREA