

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
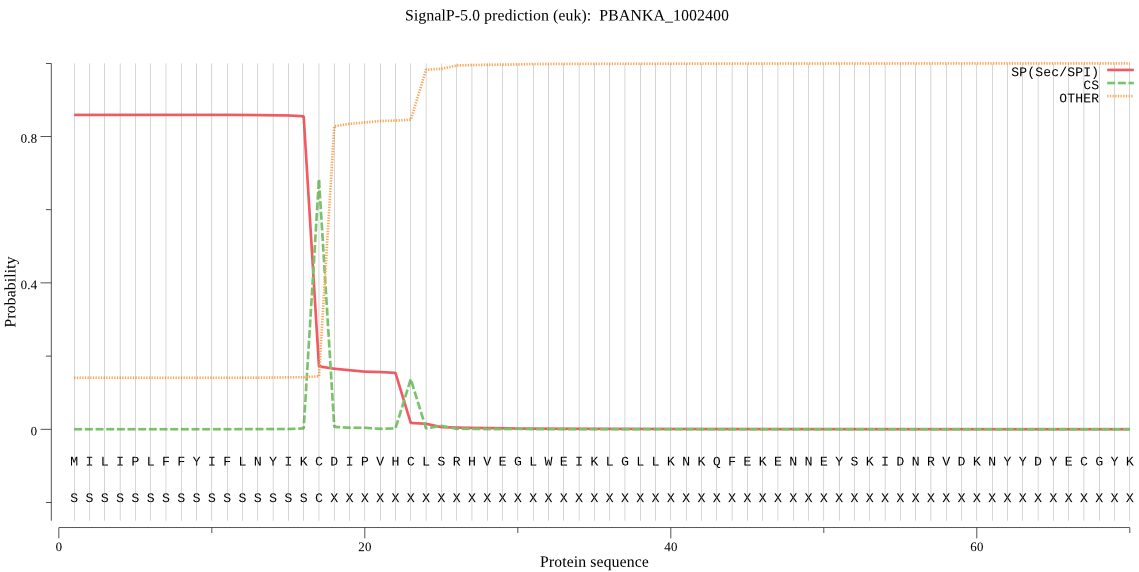
| PBANKA_1002400 | SP | 0.073564 | 0.926155 | 0.000282 | CS pos: 17-18. IKC-DI. Pr: 0.9445 |
MILIPLFFYIFLNYIKCDIPVHCLSRHVEGLWEIKLGLLKNKQFEKENNEYSKIDNRVDK NYYDYECGYKRPDDSKYHDALDPENVKKRFEIKDKKIIAFNKDRTINIIENGKSVPEYSG YWRIVYDEGLHIEIYNSKNKNKEIYFSFFKFVKNGEVSYSYCSNLVMGVVSMYHLNENTD ITKSNANIVNNSRINNCNDKITRNSMVDDINIDSSKCMNKFEENGKENNSSINNRITKKE LGEKNIIFRYAYALYGVLKNSYLNILNNTVELKENESVALNDVEKLYGNNNDVETLIDHY NDNFIDFKTMSMKRYCWSGKKLDKNSDKATNKISNMLSPLDVNADTQNHNYYINLIGDEK NKGVLTNNKGKKRKINNIQGVINNPPTKKIKENSLFNMYSNRDIALKNFDWSDENDVKER FNGNSIRIFDEAIDQKECGSCYANSASRIINSRIRIKYNYIKKIDSLSFSNEQLLICDFF NQGCNGGYIYLSLKYAYENYLYTNKCFERYENLYINKDDKNNSLCDRFDTFKIFLKKKEK NIYINNYHEKMEQKNEHIKIYNNFGNTHDNSDNRNDSNIHKKLVKGENINYIYRLVDSGK QGKGNVSDDYLNEDYILVDKTMYERGKFKLDELDSCDTKIKVTKYEYLDIENEENLKKYI YYNGPVAAAIEPSKSFIKYKKGILTGNFIKMQDGDKSNAYIWNKVDHAVVIVGWGEDTIE NLIKKKMKINNSKKITHSYDEYKSKNNDNLDENIYDKHINSYVKNTKEKNKVIKYWKVLN SWGTKWGYNGYFYILRDENYFNIRSYLLICDVNLFVKNKNVKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1002400 | 738 S | KITHSYDEY | 0.993 | unsp | PBANKA_1002400 | 738 S | KITHSYDEY | 0.993 | unsp | PBANKA_1002400 | 738 S | KITHSYDEY | 0.993 | unsp | PBANKA_1002400 | 52 S | NNEYSKIDN | 0.99 | unsp | PBANKA_1002400 | 205 S | ITRNSMVDD | 0.997 | unsp |