

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1006500 | SP | 0.026451 | 0.973425 | 0.000124 | CS pos: 23-24. FNS-YK. Pr: 0.4811 |
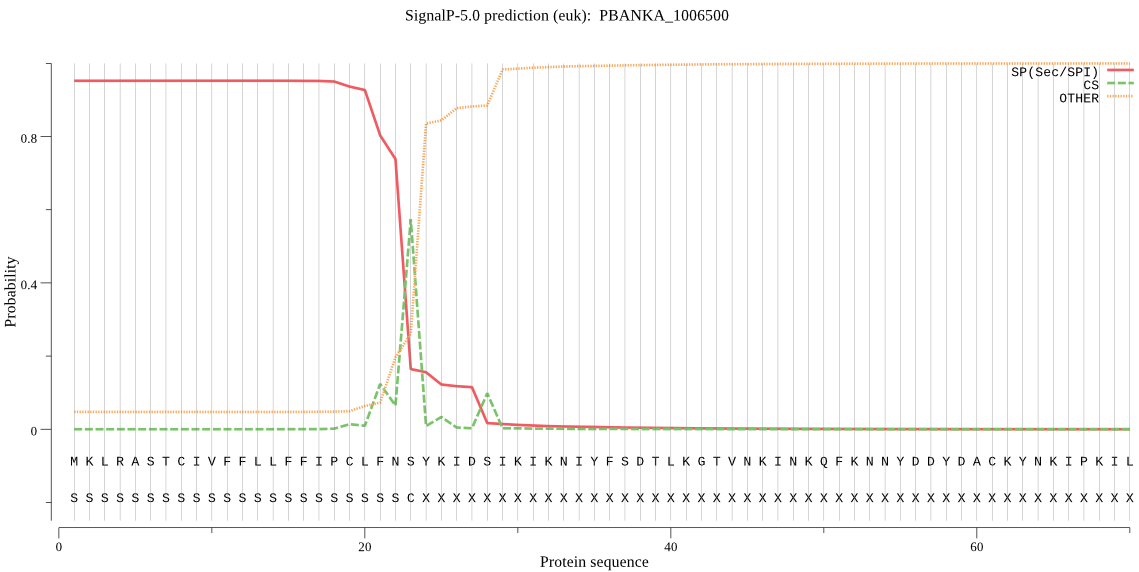
MKLRASTCIVFFLLFFIPCLFNSYKIDSIKIKNIYFSDTLKGTVNKINKQFKNNYDDYDA CKYNKIPKILIDRDKIKNNNKDSGYIVGIENTCDDTCVCIIDLQLRIIKNFIISHFKVVH KYGGVYPFFISSINSVFLKHYVNKMLEGIDASKIKCFGFSVCPGISQNMNTARNYIGDFK KRHKHIKASSINHVYAHVLSPLFFSFYNDENTFTNNYEYKNESTKKDDHKDHINMNLFDT IKKDKIQMDKINSLLKVLNNNDVTKTKLKTITTEDFLNYGSYTLKKKKIDDNNIQLNETK NNINQIKESTNMEKIPLNYIQNGYICLLVSGGSSQVYRVQKDKQNAINVCKMSQTVDISI GDIIDKVARILNLPVGLGGGPFLERESEKYIKTLNGQKNNEDISFDLFEPFPVPFASNNK INFSFSGIFNHLSKIIKELKKEKNFENEKSKYAYYCQKYIFKHLLNQLNKIMYFSELHFN IKNLFIVGGVGCNKFLFESLKILALNRNKIENQLKEYIKLKKRLKKKIKKIENNNFLAKK NLATNNMNDEIELSSSFAWNFYLKNLLKKKTSNDILLALRAFDFNSFTKLKEKGSFLLQD QFLTPSVTQPWNVYRTPINLSRDNAAMICFNTFLNIHNKINIHEDISEIKIKSTIKTQLQ NNFLLLSDIIVFDVFLDHFDHNVA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1006500 | 223 S | YKNESTKKD | 0.996 | unsp | PBANKA_1006500 | 387 S | LERESEKYI | 0.996 | unsp |