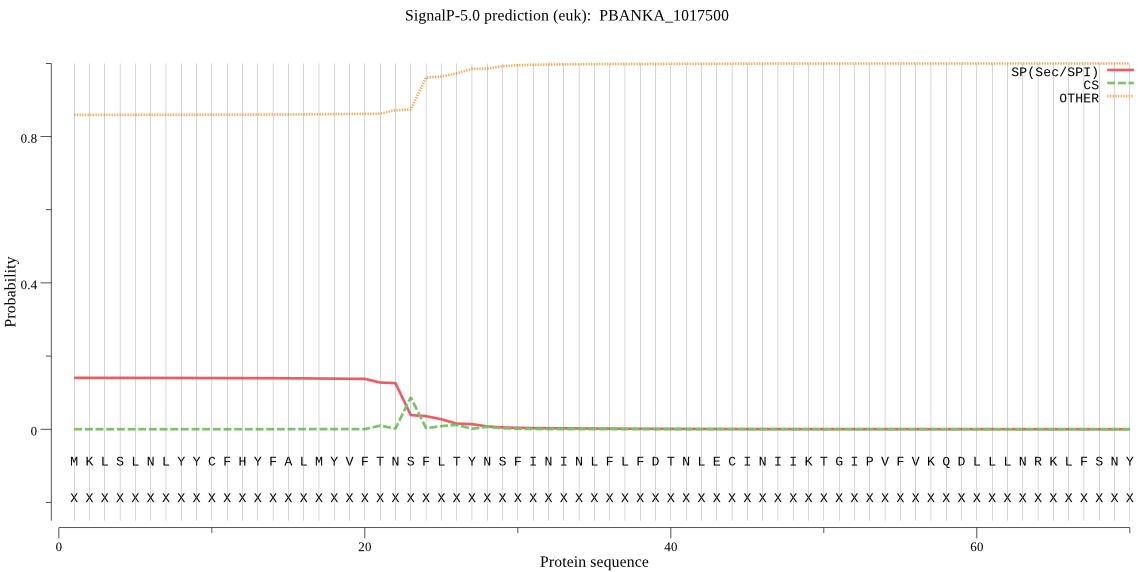
Fasta :-
MKLSLNLYYCFHYFALMYVFTNSFLTYNSFININLFLFDTNLECINIIKTGIPVFVKQDL
LLNRKLFSNYDNLDINDNTGNSENEIPTTHINIRKETENYNINCKNINDEKCIIHAEGNQ
CIKKNNNCITTLNKLKNDKTQNEQTEISDNKIIGTDITQIKEYGDSDKNLIKEKNEETIM
KKNDENVEMCKINNNENSGIELDMNKNIVDVRLNENEENCENVSTQKCTQIDDEMLKMMD
TKNILVIINEDENKNSYEEDDEEEDDDELINKHLNEVKNIIYELSFRNYEQNKPINSSDS
DITDDTSNNLDDLDENEYEFVDGVCELDDINNMHVKYKNDLEKYRSKLKENKLDEYYNDI
IKSRNLININYENEYMLKMKYHKDYNFPLHNLHIVNDKYIKNKLYKNFVLLNGDNDNLSF
QEINYYADLSKIDICPTMFKDFYLKFSLCNGKYLPLGSSFVERVCFLSYHLKQNPHLNKI
KVTKGKFILYHLFLNVVKAFKILSINKCVKHIKESNLLYIIFILNEEIIKESENEVNIGK
CLNNIIQRNKEWYEILNFTFTIMFGCSSYLKEHYYLTEEEKNNIFFKDNILDLPIFEYFN
KNIIDIFVPRNIDPKINIDLFKDISNTSGKNLKLYQTWYFCYLFIHKVYLLNKMWNIIKK
WEFSNFVPNPWYIDHIGSVLVPHIISLRHIQQNDKKFFRYIDELQMFVCFKRSKKYSIPN
YDKNEYHLVEHIVAFQGTSSPFIWLFNLIYELTPYPFLTKGKVHKAYLYIFKKAVKPYLH
ILKKKILNEIKNTKSKYSKENPYVIIFTGHSFGASMANMSSLYLENILREQGVKTKGNNN
NNDDIGNNDNDHSVYVKIYSITFGMPIFYDDKYFEDFEKSAVISNNINVDYDPIPVAMAI
PTLNGFRDPEENKKFNIIFKVEDLKKLNYDFGNIIFDGDELFNNEKNKPRSIMFLFMKYF
FKNNIFFELCEQHIYQTHYFFYFVFLTLLSRWANEAEWGTYFLIPFFIFDILSFSHKNVM
FMLKENYKKYKKLVLKKTKSNEKQEKNQVAN