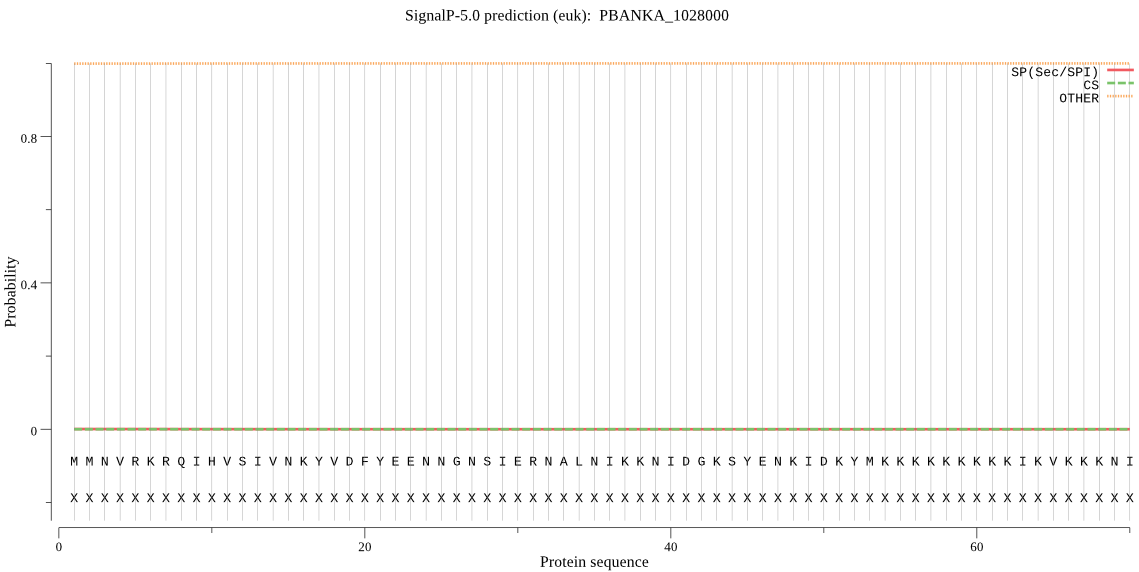
Fasta :-
MMNVRKRQIHVSIVNKYVDFYEENNGNSIERNALNIKKNIDGKSYENKIDKYMKKKKKKK
KKIKVKKKNIYINKNSVINNTEKENNNEINSSVNWSTNNSCSCLIKEDLNTTYYNKIESS
KIKKNENILLNNNDNIGYSDKENEKKVCKVKYKNKTKLFFYLIKKYPKVLFSRRSLSYYY
KLVGKYNKGILDTKKYKNERIKLRNLLYKINLRNATKRETSSEETQGLNNKTRNREINIL
DKLYVRNEFINTNRCKQKVKKIKEETEFRNDYEYISNHRYTSINNKDNTIHDDFISKIDA
KQMRIDNNSDEYYIEEKLRKYKNFQKKKKKKKEMLNKEEIFQRCDTNSLPTNYDENKKCE
MMDKIVNPLLKRDENAGVQWDFRNSKGKKLKKAIKKIEKKYIINTKRAKSGEIIFPAKYS
KNDEIKNMGNKNRDENNSEEIKKKKEPVHIPMDRYNNSNSCMYNETNYKLNAQNNKKNDN
IKIEKRDDILDNIFHPSQDMLNYKTFERIKDNYYDENDEKYVRGKYIARLEEKEMNKKNI
KTKKKSHNIDGENPKIHMNRDEYILKKKKKKKSSRKRRYLNKSGNESDNIKTKEYLILKE
IKMKQDSQKIRLNLDASCFASKGAGLYNYGQNICFFNSIIQTIVRIPYICKDLLNRLHSL
NCEKKKKKEFCFYCIFEHFGYNIISKKNVIKNTLIPYIKKYICNSYNIGYQEDVHEYLRY
FLCSLEKSSFFSSIYIQKMFTGVTKNVTICMNCNNVSLKYEQYYELSLDISSVNNLEEAL
KNFLSNEMLIGDNGYYCEKCRKKKKATKQCVINKLPRVLTIQIKRFFMNSKFNIVKNRKH
ISYPLCLDMKYYVNNYYLFKNSINDDVIHLYEKMMSNNKSNKNNKSNDNNFFNDNKSKIG
RNDYINQNKLKEENYNNVKNEINTNNFSNFEKNENNNDSIQDKYEKYNNKDLNVLREIEN
IFIQLKNEIYIKLQNRHPPISSLKNLVIEKRKEIKKELNKIKYFKFYKGAILKISKDINT
LYYCIKNSSEKNKKINLKTLVFKYRLDYYERKQKERENNKHEINFLNKKNSNNSEGFSNT
NNYEKDKFLENKGEKNLNNNKFYQNNNHFYYELTGLIKHIGSGTDYGHYIALTKSNNNIY
LQCDDNNISYVNKKDILNCAKNAYVFVYTCTHPQFIDFYNAYVDVLEKKKFDINLPVFEK
RVEFRERITMPKKKFISRSLCYAIRG