

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
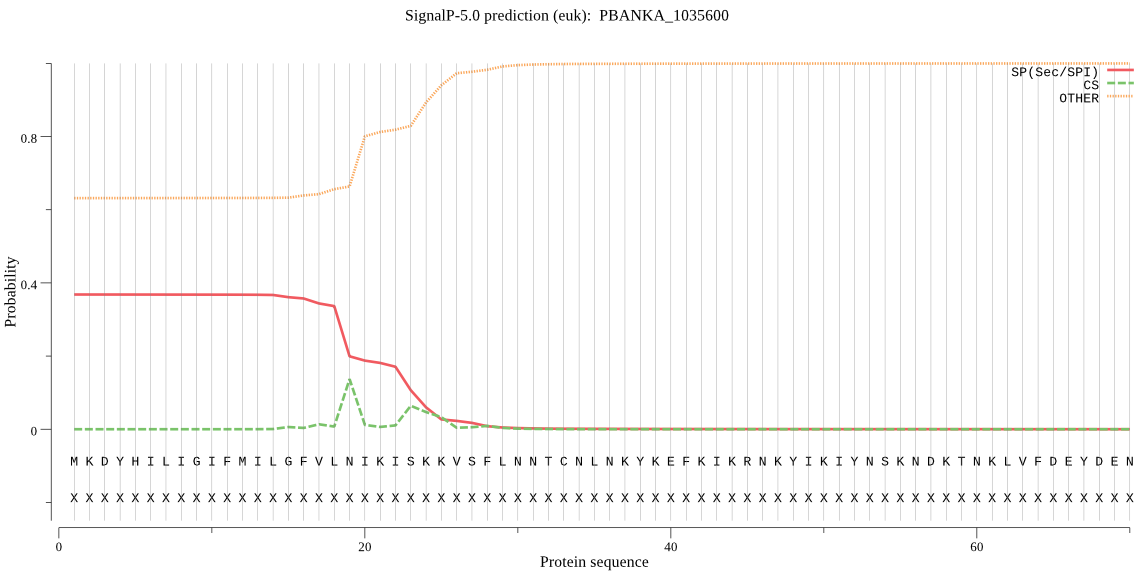
| PBANKA_1035600 | SP | 0.414369 | 0.566504 | 0.019127 | CS pos: 25-26. SKK-VS. Pr: 0.2550 |
MKDYHILIGIFMILGFVLNIKISKKVSFLNNTCNLNKYKEFKIKRNKYIKIYNSKNDKTN KLVFDEYDENCIKALIMAREVAKNNNENEVLLKHLLVAIIKTDSHIIDHIFKYFNISLQH FINKFNTEIAKNNFKKIGQEPHDDNHIQPQQIELQKCSTENNDQEKKFINNIIDKKIKNE EYNNVVLNKLDKESTKDNNTELTSNENYESEKITDILSNYNNNNKMEKNDIDTESITHQN EEEKQNDEIFLVNNNSVKRNSQKELKLNKEIKFSENCKLVLQNAILEAKKKKKIIVNLTD ILISMINIAQENKECDFVKYLNELNISLSDLKKQVNNYDENNLNSNLIHYSIIKNTIQNH GENNNEHQVGNKNLNHYSDKQNRNMPDQSNNIINNMNNEYINQEKYFKNNDDNNFTQNNK FISSIITKDCLIDMIQRAFEKRLDNFFGRKDEIKRIIEILGRKKKSNPLLIGEAGVGKTA IVEQLAYLILKNKVPYHLQNCKIFQLNLGNIVAGTKYRGEFEEKMKQILANISKDKKNIL FIDEIHVIVGAGSGEGSLDASNILKPFLSSDNLQCIGTTTFQEYSKYIETDKALRRRFNC VNIKPLNSNETYKLLKKIKNSYEQYHNIYYTNEALKSIILLSDDYLPSLNFPDKAIDILD EAGSYQKIKYERYITKKLRNENNIKIDPIKVKENVNTNKTDHTINTTSNKIDNNSTNITS KIESNYLIDDLHMKYVTSDVIENIVSKKSSISFIKKNKKEEEKIQNLKEKLNKIIIGQEK VIDILSKYLFKAITNIKDANKPIGTLLLCGSSGVGKTLCAQVISQYLFNDDNIIVINMSE YIDKHSVSKLFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHSDVLHVLLQILDN GLLTDSKGNKVSFKNTFIFMTTNVGSDIITDYFKVYNKNYANFGFKYYTQKDKIEKAELQ NPTNSNTHNDKDKIQTDLDIFEEKIKTNKWYDELQPEIEEELKKKFLPEFLNRIDEKILF RQFLKRDIIQILENMIEDLKKRIKKRKKLNILIDDNIIKYICSDENNIYDINFGARSIRR ALYKYIEDPIAAFLISNEYDPNDTIHLTLLPNNKIHVKLLKHTITSLS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1035600 | 194 S | LDKESTKDN | 0.997 | unsp | PBANKA_1035600 | 261 S | VKRNSQKEL | 0.998 | unsp |