

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1107100 | SP | 0.059532 | 0.937491 | 0.002977 | CS pos: 20-21. IPA-HN. Pr: 0.3780 |
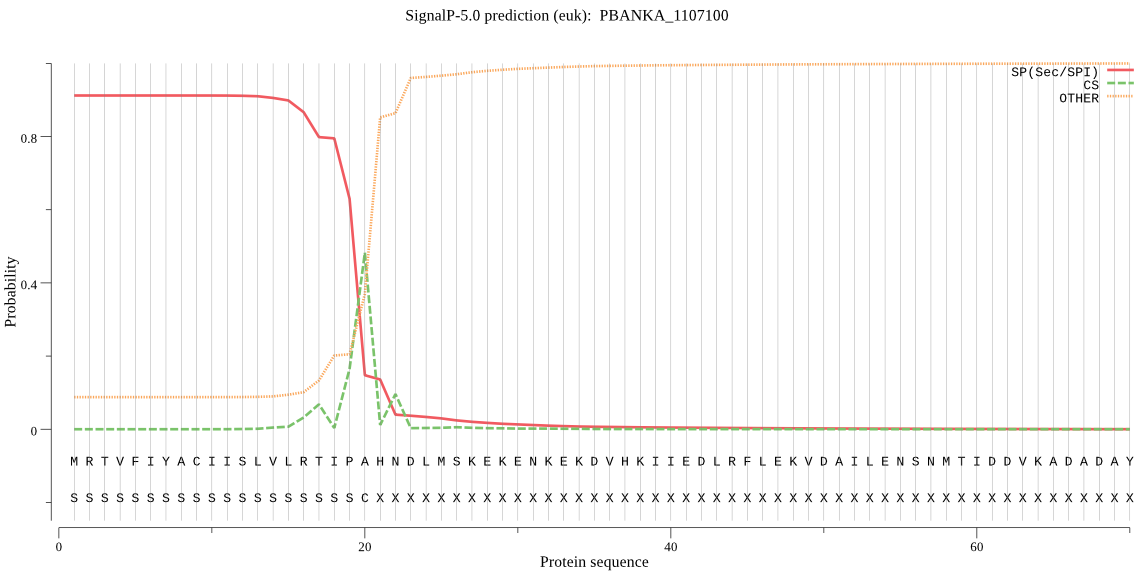
MRTVFIYACIISLVLRTIPAHNDLMSKEKENKEKDVHKIIEDLRFLEKVDAILENSNMTI DDVKADADAYNPDEDAPKEELNKIEMEKKKAEEEAKNSKKKILERYLLDEKKKKSLRLIV SENHATSPSFFEESLIQEDFMSFIQSKGEIVNLKNLKSMIIELNSDMTDKELEAYITLLK KKGAHVESDELVGADSIYVDIIKDAVKRGDTSINFKKMQSNMLEVENKTYEKLNNNLKKS KNSYKKSFFNDEYRNLQWGLDLARLDDAQEMITTNSVETTKICVIDSGIDYNHPDLKGNI YVNLNELNGKEGIDDDNNGIIDDIYGVNYVNNTGDPWDDHNHGSHVSGIISAIGNNSIGV VGVNPSSKLVICKALDDKKLGRLGNIFKCIDYCINKKVNIINGSFSFDEYSTIFSSTIEY LARLGILFVVSSSNCSHPPSSIPDITRCDLSVNSKYPSVLSTQYDNMVVVANLKKKINGE YDISINSFYSDIYCQVSAPGANIYSTASRGSYMELSGTSMAAPHVAGIASIILSINPDLT YKQVVNILKNSVVKLSSHKNKIAWGGYIDILNAVKNAISSKNSYIRFQGIRMWKSKKRN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1107100 | 511 S | ASRGSYMEL | 0.997 | unsp | PBANKA_1107100 | 511 S | ASRGSYMEL | 0.997 | unsp | PBANKA_1107100 | 511 S | ASRGSYMEL | 0.997 | unsp | PBANKA_1107100 | 127 S | NHATSPSFF | 0.992 | unsp | PBANKA_1107100 | 243 S | KSKNSYKKS | 0.997 | unsp |