

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
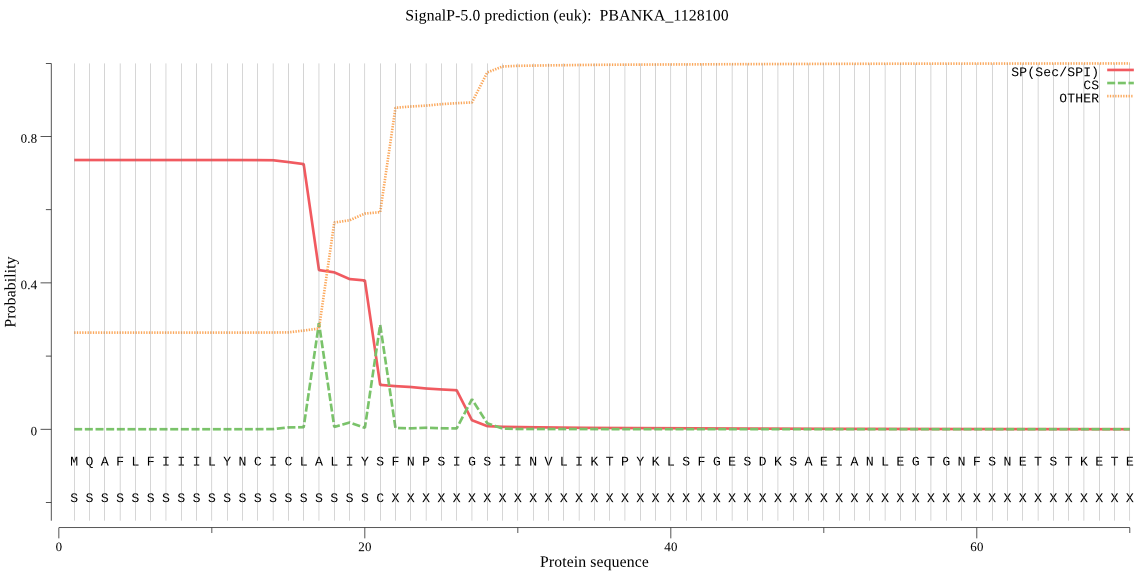
| PBANKA_1128100 | SP | 0.038524 | 0.961167 | 0.000309 | CS pos: 21-22. IYS-FN. Pr: 0.5592 |
MQAFLFIIILYNCICLALIYSFNPSIGSIINVLIKTPYKLSFGESDKSAEIANLEGTGNF SNETSTKETEQIVEFESKESKNDDKYVLNNESKPESVVDYEVAATKEAIVEEASIDNETI IEETVNKDDPSDEIKRIKEVTAEEIEKLKKSAAKEIKKIKKAAAGEVEKIKETVAEEIEI AKEIIAGSVLEKKEKEKEYVSEHTEENKEKGPISEHTEENKEKESVSEHTEESKEDTLKE IEVSKNVNIENETIVKEIDKSNDEMLSETEKEYNTDEMIQEKYGEFKIGENSYYWENAYI DAEGTVDDGMEIVGSSKRSNSAISDKKIKNSVYLIPGLGGSTLIAEYNDADIESCSSKLL HSKPYRIWLSISRLFSIRSNVYCLFDTLKLDYDRENKIYRNKPGVIINVENYGYIKGVAF LDYVKNKPLRLTRYYGILADKFLENEYIDGKDILSAPYDWRFPLSQQKYNVLKSHIEHIY KIKQEIKVNLIGHSLGGLFINYFLSQFVDEEWKKKHINIVIHINVPFAGSIKAIRALLYN NKDYTLFKLKNILKVSISGSLMRAISHNMGSPLDLLPYRKYYDRDQIVVIINMDEFPIDE KLAQSIVTECGIYNESCYTDREDVNLKTYTLSNWYELLSNDMREKYENYIQYTDRFFSVD HGIPTYCLYSTTKKKNTEYMLFYQDTHFIQDPIIYYGIGDGTIPLESLEACKKLQNVKEA KHFEYYKHIGILKNDIVSDYIYNIIDKNTTN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1128100 | 96 S | SKPESVVDY | 0.997 | unsp | PBANKA_1128100 | 96 S | SKPESVVDY | 0.997 | unsp | PBANKA_1128100 | 96 S | SKPESVVDY | 0.997 | unsp | PBANKA_1128100 | 225 S | KEKESVSEH | 0.994 | unsp | PBANKA_1128100 | 233 S | HTEESKEDT | 0.997 | unsp | PBANKA_1128100 | 321 S | KRSNSAISD | 0.994 | unsp | PBANKA_1128100 | 324 S | NSAISDKKI | 0.995 | unsp | PBANKA_1128100 | 376 S | SRLFSIRSN | 0.995 | unsp | PBANKA_1128100 | 65 S | SNETSTKET | 0.998 | unsp | PBANKA_1128100 | 80 S | ESKESKNDD | 0.996 | unsp |