

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1134600 | SP | 0.010267 | 0.989386 | 0.000348 | CS pos: 16-17. ASG-NS. Pr: 0.5152 |
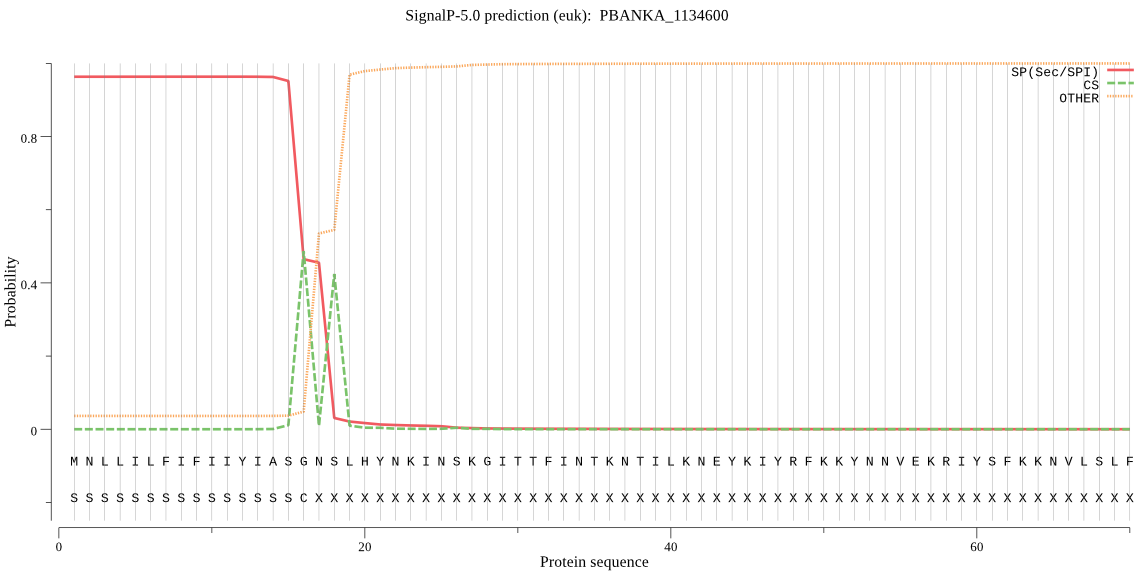
MNLLILFIFIIYIASGNSLHYNKINSKGITTFINTKNTILKNEYKIYRFKKYNNVEKRIY SFKKNVLSLFNTLKNKEKITNLLENISNNVKIKFPNRFIYYNYLFNKCKLDRILIVINTL LYLYLNRVDKNEEKKIFFTKGNLVQIKDEQKAAKYQCNYYDIYKNKNYKTLFSSIFIHKN ILHLYFNMSSLMSIYKIISPIYSNSQILITYLLSGFLSNLISYIYYIKPPKKNIFLKDII DQNYYSRNIPLNKPNKIICGSSSAIYSLYGMYITHMIFFYFKNNYIANTSFLYNIFYSFL SSLLLENVSHFNHILGFMCGFFMSSTLILFDNN