

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1316000 | SP | 0.154896 | 0.844465 | 0.000639 | CS pos: 18-19. IIG-IK. Pr: 0.4124 |
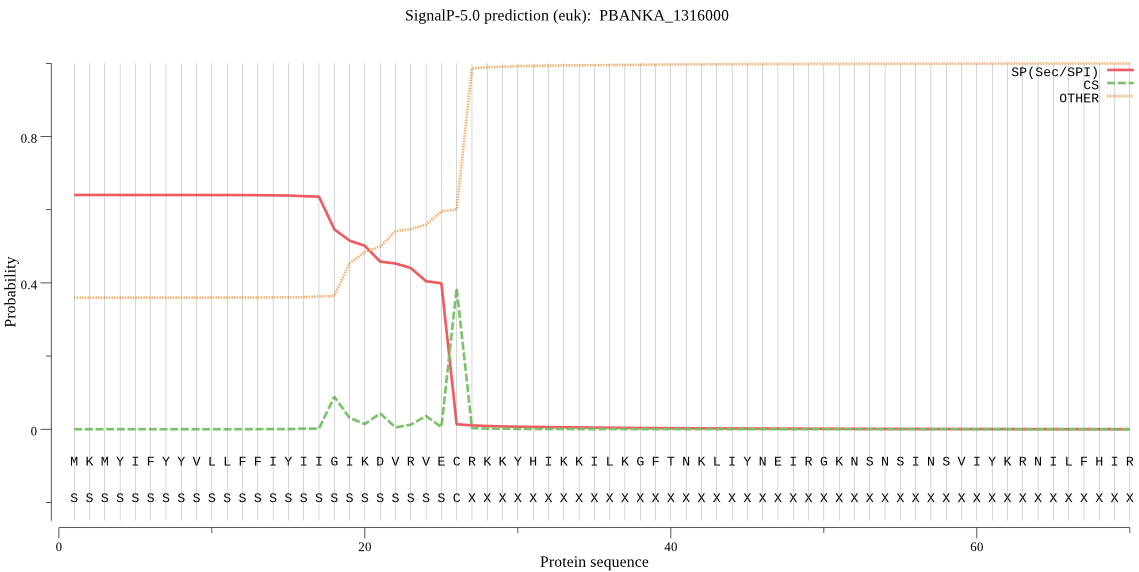
MKMYIFYYVLLFFIYIIGIKDVRVECRKKYHIKKILKGFTNKLIYNEIRGKNSNSINSVI YKRNILFHIRIPKKTLQGRNLFSNDIRNKKINEFLQKKKNSCYLHINSNVNEIKKKRDRI LKNVFINAKLLQEYFNNFKSSLLYKYKNTKIITKLFLSTSFLVMILNVFGLRPEDIALHA KRIIRAFEFYRIITSALFYGDISLYVLTNIYMLYLQSQELEKSVGSSETLAFYLSQITIL SAICSYIKKPFYSTALLKSLLFTNCMLNPYNKSNLIFGINIYNMYLPYLSIVIDILHAQD FKASLSGILGIISGYIYYISNIYLLEKCNKKFFKIPQILRNYLDSFNTDEFVF