

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
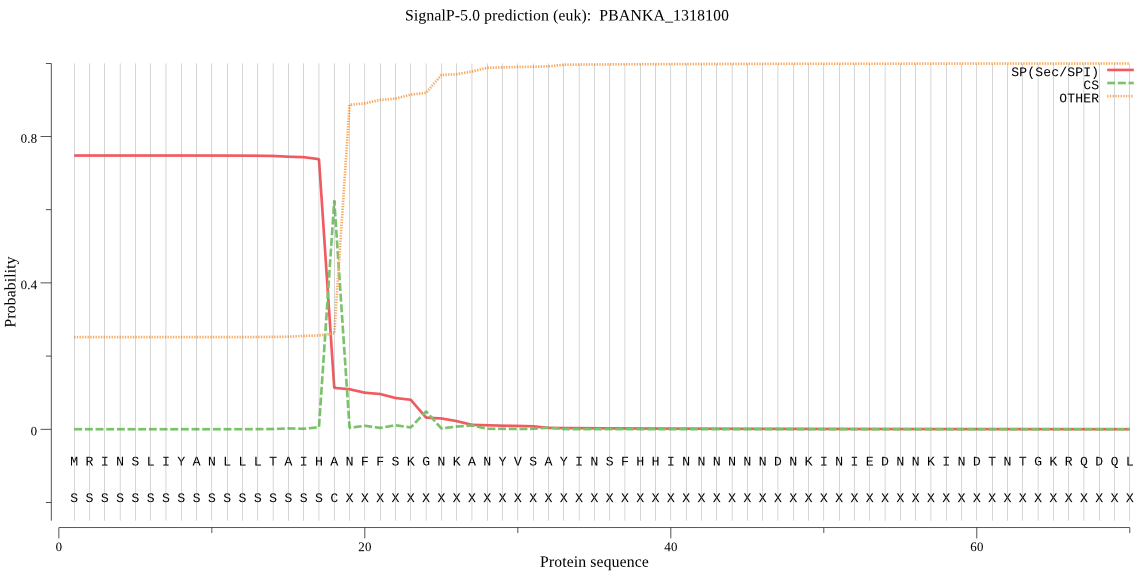
| PBANKA_1318100 | SP | 0.048859 | 0.925408 | 0.025733 | CS pos: 18-19. IHA-NF. Pr: 0.9545 |
MRINSLIYANLLLTAIHANFFSKGNKANYVSAYINSFHHINNNNNNDNKINIEDNNKIND TNTGKRQDQLNNFFTRRFYTFHNKNQYLREEKRNSKEVNLTNNNTNSYNMESSNEQNNQM MGGDSYKERLQNLKKYMAEHNIDVYIIINSDAHNSEIINDQDKKIYYLTNYSGADGILIL TKDAQIIYVNALYELQANKELDTKFFTLKVGRITNRDEIFQTIADLKFNTIAFDGKNTSV SFYEKLKDKIKIQFPDKKIQEKFIYKNSINQVVKDNNINLYVLESPLVTVPNSDVNKKPI FIYSREFGGSCAAQKIQESFDFFIENPDVDSLLLSELDEIAYLLNLRGYDYKYSPLFYSY VYLKYNRDKGIIDDIILFTKVENVQKNVLAHLERIHVKLMDYDSVVSYLTNNVSSKSENT KNNNGKNIILGSVHENRSPRYDISLSPHINLMIYMLFNKEKVLLKKSPIADMKAVKNYVE IDSIKEAHVLDGLALLQFFHWCEEKRKTKELFKETEISLRDKIDYFRSTKKNFIFPSFST ISAIGPNSAVIHYESTEDTNAKITPNIYLLDSGGQYLHGTTDVTRTTHFGEPTPEEKKLY TLVLKGHLSLRKVIFASYTNSMALDFLARQPLFNNFLDYNHGTGHGVGTCLNVHEGGCSI SPATGTPLKENMVLSNEPGYYWADHFGIRIENMQYVVTKKQTNDAKFLTFNDLTLYPYEK KLLDYYLLTPEEIADINEYHQTIRNTLLPRIKENPSDYAKGIDQYLMDITEPIHTK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1318100 | 415 S | NNVSSKSEN | 0.994 | unsp | PBANKA_1318100 | 415 S | NNVSSKSEN | 0.994 | unsp | PBANKA_1318100 | 415 S | NNVSSKSEN | 0.994 | unsp | PBANKA_1318100 | 483 S | VEIDSIKEA | 0.993 | unsp | PBANKA_1318100 | 518 S | ETEISLRDK | 0.99 | unsp | PBANKA_1318100 | 555 S | IHYESTEDT | 0.995 | unsp | PBANKA_1318100 | 95 S | EKRNSKEVN | 0.996 | unsp | PBANKA_1318100 | 125 S | MGGDSYKER | 0.995 | unsp |