

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
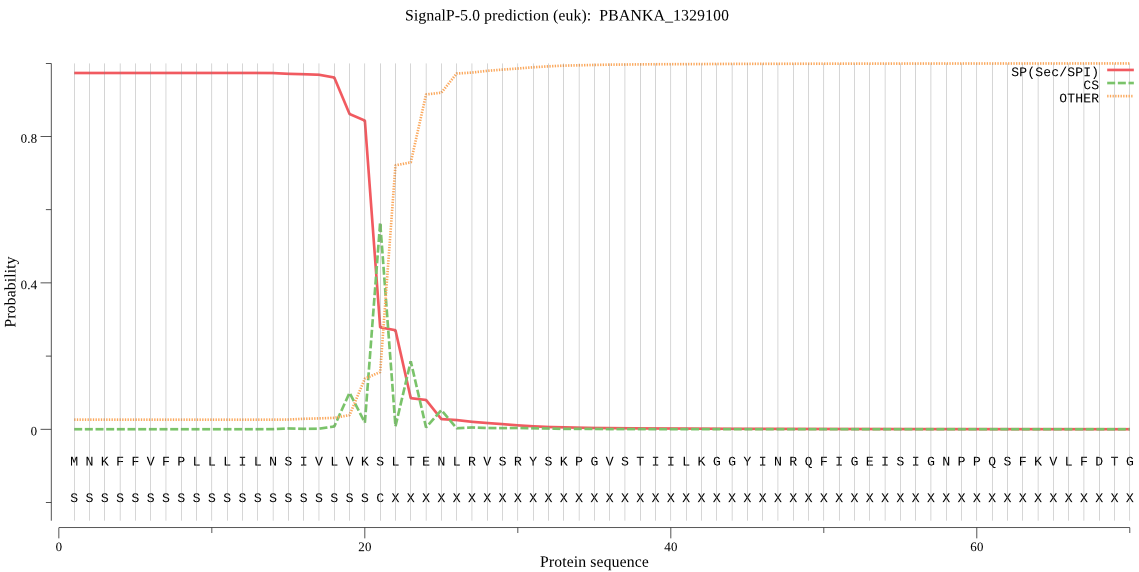
| PBANKA_1329100 | SP | 0.002623 | 0.996842 | 0.000535 | CS pos: 21-22. VKS-LT. Pr: 0.7386 |
MNKFFVFPLLLILNSIVLVKSLTENLRVSRYSKPGVSTIILKGGYINRQFIGEISIGNPP QSFKVLFDTGSTNLWIPSKNCYAKACYNKKKYDYNISKNYRISSSKNPVNIFFGTGKVQI AYATDDIHLGSIKVRNQEFGIANYMSDDPFSDMQFDGLFGLGISEDIKRKGLIYDNIPRN SSRKNVFSIYYPKSVDDNGAITFGGYDKKYIEPNSNIDWFVVSSRKYWTIKMTGIKINGL FLEVCSGNIEGYCDAVIDTGTSSIAGPQNDLILLTKLLNPVKSCQNKTLLKNFSFVFSDE NGVEKEYELTSNDYIVNSFKVDPILKNPCNFAFMPINISSPNGYLYILGQVFLQKYYAIF EKDNMRIGLAKSI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1329100 | 182 S | PRNSSRKNV | 0.995 | unsp | PBANKA_1329100 | 182 S | PRNSSRKNV | 0.995 | unsp | PBANKA_1329100 | 182 S | PRNSSRKNV | 0.995 | unsp | PBANKA_1329100 | 194 S | YYPKSVDDN | 0.991 | unsp | PBANKA_1329100 | 29 S | NLRVSRYSK | 0.991 | unsp | PBANKA_1329100 | 104 S | YRISSSKNP | 0.992 | unsp |