

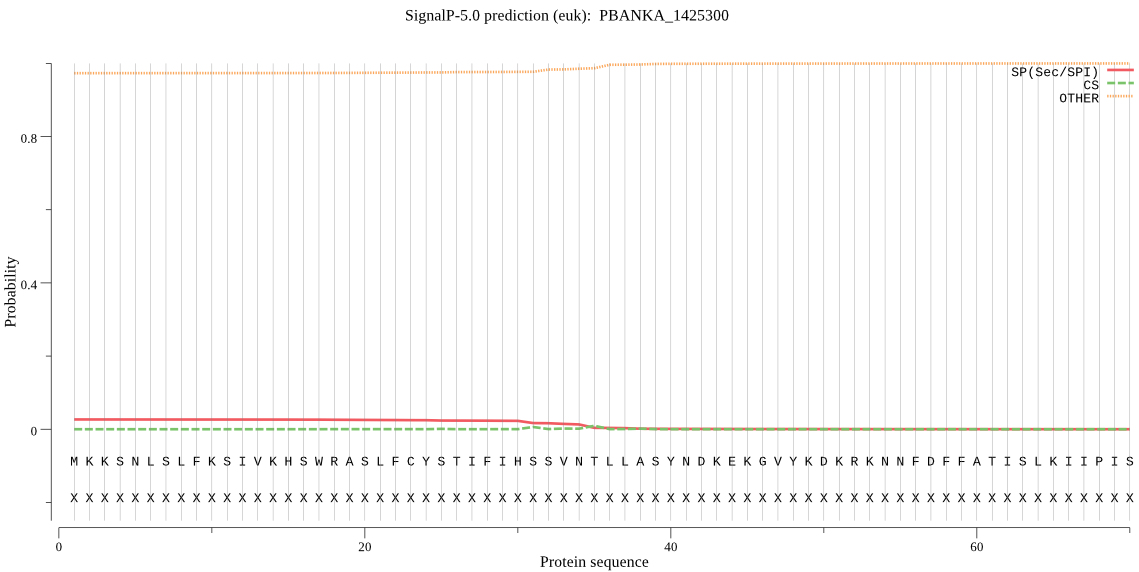
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1425300 | OTHER | 0.903496 | 0.027910 | 0.068595 |
MKKSNLSLFKSIVKHSWRASLFCYSTIFIHSSVNTLLASYNDKEKGVYKDKRKNNFDFFA TISLKIIPISYCSCQLVYKCDNNNALPTETEYNNLELFKRVFFKLIQYEYFIIHKIIRCI NEFIIFVLYSLANFYLYFHSIFFNICNLLSKIYHGESNNGSLHAYPNIPYNIINSFVTIH KIYEHNKIPLKHDFKNDQLEYMGSEFIYDKRGYILTAAHNITNLEDKFVVKNNNGLYFAT LLGLHKGSDVCVMKINSEEPFSYISLDKIRDELKQGESVVTYGQIQDFDKETYSMSGSPL LDQYGNLVGMIQKKIDNYGLALPANILKNVAIHLQNKGVYKEPFLGIVFREKALIIQNSR SLTKELKISSILANSSADLGNLKKEDIISKINNKDIESICDVHEIINSTSDRYINVDGIR NDKKFKTQIKLW
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PBANKA_1425300 | 16 S | IVKHSWRAS | 0.994 | unsp |