

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1425350 | SP | 0.451328 | 0.525158 | 0.023514 | CS pos: 27-28. TWC-FI. Pr: 0.5339 |
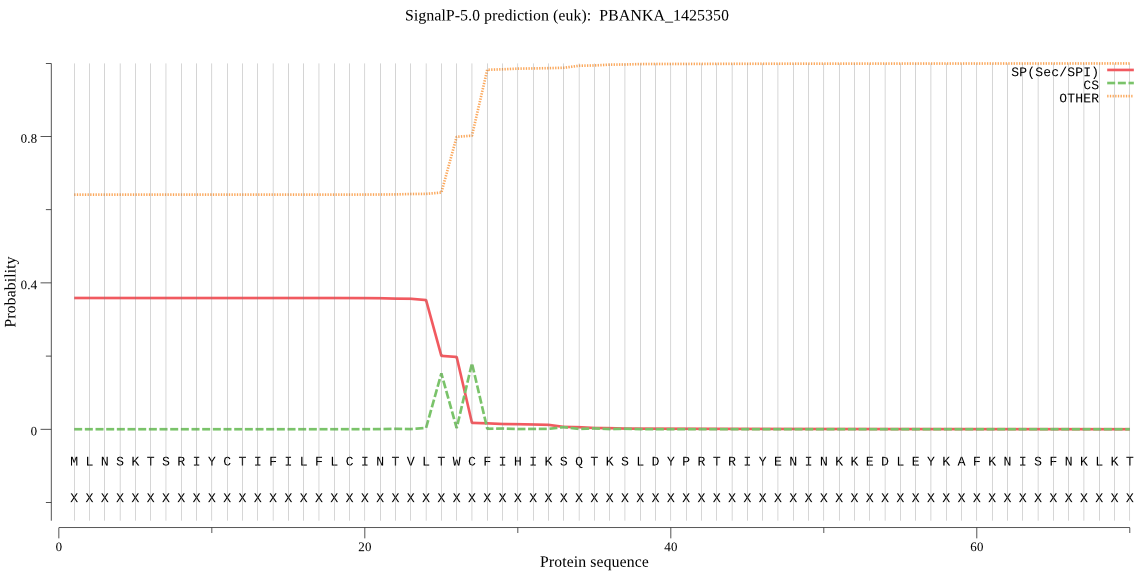
MLNSKTSRIYCTIFILFLCINTVLTWCFIHIKSQTKSLDYPRTRIYENINKKEDLEYKAF KNISFNKLKTSLKNFLVLKKKINISNVKYVSFLGKYYNYEKIDNYGVNEICILGRSNVGK STFLRNFIKYLINVNDNKNIKVSKRSGCTRSINLYSFENAKKKKLFILTDMPGLGYAEGI GEKKMEYLKKNLDDYVYLRNQICLFFILIDMSVDIQKIDISLMDAIRKTSIPLRVICTKC DKFNGNVNHRLNGIKTFYKLEKTPIHISKFSNNNYINIFKEIQYHCNLDNS