

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
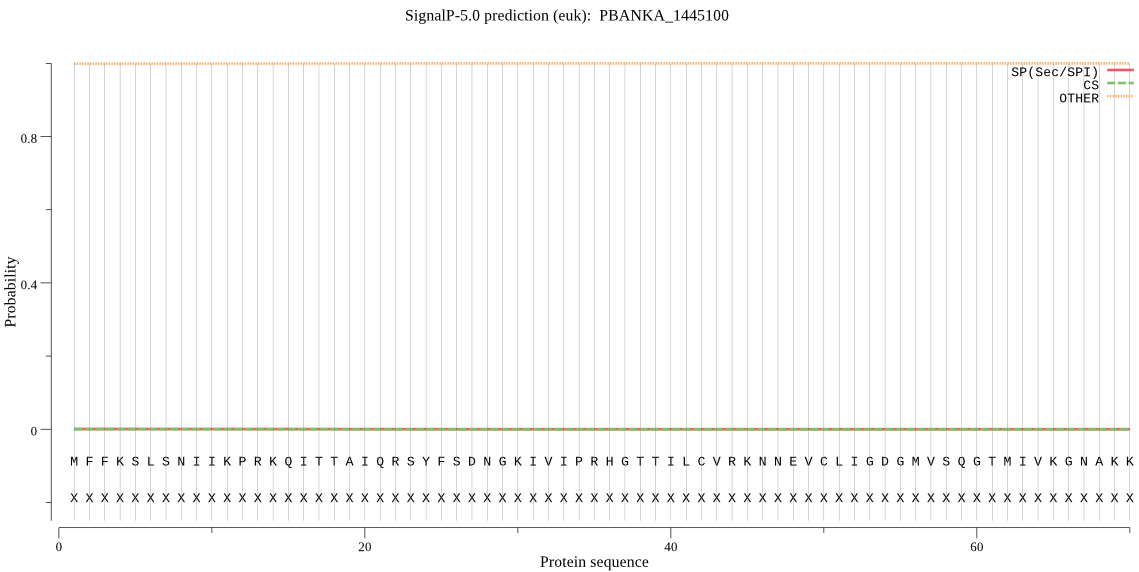
| PBANKA_1445100 | mTP | 0.384151 | 0.000209 | 0.615640 | CS pos: 24-25. RSY-FS. Pr: 0.4606 |
MFFKSLSNIIKPRKQITTAIQRSYFSDNGKIVIPRHGTTILCVRKNNEVCLIGDGMVSQG TMIVKGNAKKIRRLKDNILMGFAGATADCFTLLDKFETKIDEYPNQLLRSCVELAKLWRT DRYLRHLEAVLIVADKDTLLEVTGNGDVLEPSGNVLGTGSGGPYAIAAARALYDIENLSA KDIAYKAMNIAADMCCHTNHNFICETL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PBANKA_1445100 | 179 S | IENLSAKDI | 0.996 | unsp |