|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
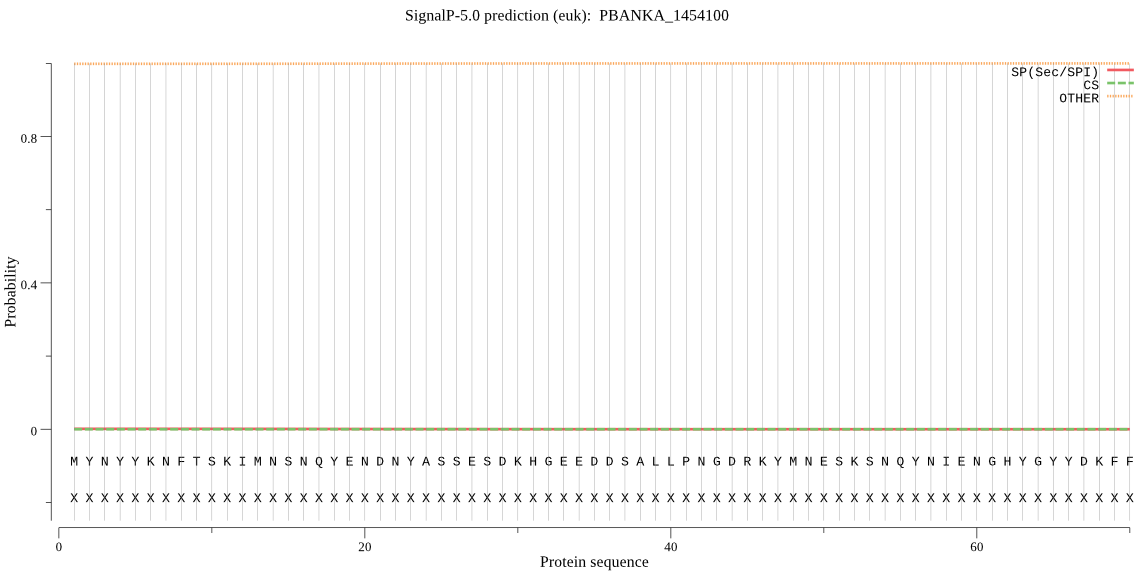
PBANKA_1454100 | OTHER | 0.999750 | 0.000008 | 0.000241 | |
No Results
-
Fasta :-
>PBANKA_1454100
MYNYYKNFTSKIMNSNQYENDNYASSESDKHGEEDDSALLPNGDRKYMNESKSNQYNIEN
GHYGYYDKFFGKRRNKKWNYFVIFFLGVCLGFAIWPFLMTIITYKLVYKDNYNNHNFPTS
DNLNSLKPYVNDKNRKNDNPNNRAPPSDHGQKKPSPHFKPVKFDEIAGIDESKLELLEVV
DFIKNRERYQEMGARMPKGVLLVGPPGSGKTMLARAVATEANVPYIYTSGPEFIEIYVGQ
GAKRIRQLFSHARSVAPSIVFIDEIDAIGGKRSSSASNGAGQKEHDQTLNQLLVEMDGFS
NTVHIMVIGATNRIDTLDSALLRPGRFDRIVYVPLPDVNGRKKILEIYIKKIKSNLNAND
IERMSRLTPGFSGADLENLVNEATILATRNKKSLVTINELFEARDKVSMGPERKSLKQSE
HQRRITAYHEAGHAIVAYFLHPKTDPIHKATIISRGNALGYVEQIPVDDRHNYFKSQMEA
KLAVCMGGRTAEEIVFGKSETSSGASSDISRATDIAYKMVTEWGMSDKLGPLNYKKRMGD
GYSSMRLSAQTISAIETEVKTLVEKGKGISEEILRRHRKELDNLAFALLDKETLTGDEIK
KIVDPNNSRDQSNRYQLLNKDSKNDDTQNDHNNSGKKSVRNDGKTNTSKNEKNEPNSHEN
KHKDSREIDTNDKDTTNNMKSDEVNDTNKMENFIESKYIDTKDITTLSSENEIGILSKHF
KNENKKLKKKYSRDPNTKNCNVLKMVNSHRPSYYSKNENFDSENSINNSLDDYSNNFRNT
RDSENNYDNNNPVKNTSEPFTNDFSEENNNDNLSSVKTKDINYNNFQEIVDISLIKNVSE
MKKFNIYEHNLNKLFLFDIFNN
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PBANKA_1454100.fa
Sequence name : PBANKA_1454100
Sequence length : 862
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.315
CoefTot : -1.321
ChDiff : 3
ZoneTo : 18
KR : 2
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.518 2.412 0.472 0.941
MesoH : -0.558 0.294 -0.342 0.207
MuHd_075 : 20.452 7.947 6.318 4.174
MuHd_095 : 27.582 19.176 8.003 6.081
MuHd_100 : 26.721 19.188 8.374 6.267
MuHd_105 : 21.822 15.983 7.475 5.658
Hmax_075 : 2.450 -0.400 -2.660 2.130
Hmax_095 : 7.525 3.763 -1.026 3.124
Hmax_100 : 6.500 2.900 -1.042 2.930
Hmax_105 : 10.600 5.100 0.560 3.530
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9467 0.0533
DFMC : 0.9594 0.0406
-
Fasta :-
>PBANKA_1454100
ATGTATAACTATTACAAAAATTTTACAAGTAAAATTATGAATTCTAATCAATATGAAAAT
GACAATTATGCATCTTCCGAATCAGACAAGCATGGTGAAGAAGATGATTCTGCATTACTT
CCAAATGGAGATAGAAAATATATGAATGAGAGCAAGAGTAATCAATATAATATTGAGAAT
GGGCATTATGGATATTATGATAAATTTTTTGGTAAACGAAGAAACAAAAAATGGAATTAT
TTTGTCATATTTTTTCTAGGCGTATGCCTAGGATTCGCTATATGGCCATTTCTTATGACA
ATCATTACATATAAATTAGTTTATAAAGATAATTATAATAATCATAATTTTCCAACAAGT
GATAATTTGAATTCTTTAAAACCTTATGTAAATGATAAAAATAGAAAGAATGATAATCCT
AATAATAGAGCGCCTCCAAGCGACCACGGTCAAAAAAAGCCATCTCCACATTTTAAACCC
GTAAAATTTGATGAAATTGCTGGAATAGATGAATCAAAATTAGAACTTTTAGAAGTTGTT
GATTTTATAAAAAATAGAGAACGCTATCAAGAAATGGGTGCACGAATGCCTAAAGGTGTG
TTATTAGTTGGACCTCCAGGATCTGGTAAAACTATGCTAGCTCGAGCAGTAGCTACCGAA
GCTAATGTTCCATATATATATACATCTGGTCCAGAATTTATAGAAATATATGTAGGTCAA
GGAGCAAAAAGAATCAGGCAATTATTTTCACATGCTAGATCAGTTGCACCGTCAATTGTT
TTTATCGATGAAATTGATGCAATAGGAGGGAAACGTAGTTCTAGCGCATCTAATGGTGCA
GGGCAAAAAGAACATGATCAAACATTAAATCAATTATTAGTTGAAATGGATGGATTTAGT
AATACTGTGCATATTATGGTCATAGGGGCAACGAATAGAATTGATACTTTAGATAGTGCT
TTACTTAGACCTGGAAGATTTGATAGAATTGTATATGTTCCATTACCAGATGTAAACGGC
AGGAAAAAAATACTAGAAATTTACATAAAAAAAATTAAAAGTAATTTAAATGCGAATGAT
ATTGAAAGAATGTCGAGATTGACACCTGGTTTTTCGGGCGCCGATTTAGAAAACCTTGTA
AATGAAGCAACTATATTAGCTACGCGAAACAAAAAAAGTTTAGTTACTATAAACGAATTA
TTTGAAGCACGAGATAAAGTTTCGATGGGGCCTGAAAGAAAATCGTTAAAACAATCAGAA
CATCAAAGGAGAATAACTGCTTATCATGAAGCTGGGCATGCAATAGTTGCATATTTTCTT
CACCCCAAAACAGATCCTATACATAAAGCTACTATTATTTCTAGAGGTAATGCTTTAGGT
TATGTAGAGCAAATACCTGTTGATGATAGGCATAATTATTTTAAAAGTCAAATGGAAGCA
AAATTAGCAGTATGTATGGGGGGAAGAACAGCAGAAGAAATTGTTTTCGGGAAATCTGAA
ACAAGTAGTGGAGCATCTAGTGATATTTCAAGAGCTACTGATATAGCTTATAAAATGGTA
ACCGAGTGGGGAATGTCAGATAAATTAGGTCCATTAAATTATAAAAAAAGAATGGGTGAT
GGGTATTCTTCTATGAGATTATCAGCTCAAACTATATCAGCTATAGAAACGGAAGTTAAA
ACTTTAGTAGAAAAGGGAAAAGGCATATCAGAAGAAATTTTAAGAAGACATAGAAAAGAA
TTAGATAATTTAGCCTTTGCATTATTAGATAAAGAAACATTAACTGGAGACGAAATTAAA
AAAATAGTTGATCCTAATAATTCAAGGGATCAATCAAATAGATATCAACTCTTGAATAAA
GATTCTAAGAATGATGATACACAAAATGATCACAATAATAGTGGAAAGAAATCTGTACGA
AACGATGGGAAAACAAACACATCAAAAAACGAAAAAAATGAACCAAATTCTCATGAAAAT
AAGCATAAAGATTCTAGAGAAATTGATACCAATGACAAAGATACCACCAATAATATGAAA
TCTGATGAAGTTAATGATACTAATAAAATGGAGAATTTTATCGAAAGTAAATACATTGAT
ACTAAAGATATAACGACACTTTCTTCTGAAAACGAAATTGGAATTTTAAGTAAACATTTT
AAAAATGAAAACAAAAAACTTAAGAAAAAATATTCGAGGGATCCAAACACCAAGAATTGC
AATGTGCTAAAAATGGTAAATAGTCACAGGCCATCATATTATTCTAAAAATGAAAACTTT
GACTCAGAAAATAGTATTAATAACTCATTGGATGATTATTCTAACAATTTTAGAAATACT
AGAGATAGCGAAAACAACTATGATAATAATAACCCTGTAAAAAATACTAGTGAACCTTTT
ACAAACGATTTTTCTGAAGAAAATAATAATGATAATTTAAGTTCTGTAAAAACAAAAGAT
ATAAATTATAATAATTTTCAGGAAATAGTAGATATATCACTTATTAAAAATGTTTCTGAA
ATGAAGAAGTTCAATATTTATGAGCATAATTTGAATAAATTATTTTTATTTGACATTTTT
AATAATTAA
- Download Fasta
- title: ATP binding site
- coordinates: P205,P206,G207,S208,G209,K210,T211,M212,D263,N312
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
PBANKA_1454100 | 275 S | KRSSSASNG | 0.995 | unsp | PBANKA_1454100 | 275 S | KRSSSASNG | 0.995 | unsp | PBANKA_1454100 | 275 S | KRSSSASNG | 0.995 | unsp | PBANKA_1454100 | 415 S | PERKSLKQS | 0.997 | unsp | PBANKA_1454100 | 657 S | NEPNSHENK | 0.995 | unsp | PBANKA_1454100 | 681 S | NNMKSDEVN | 0.991 | unsp | PBANKA_1454100 | 783 S | NTRDSENNY | 0.997 | unsp | PBANKA_1454100 | 28 S | ASSESDKHG | 0.992 | unsp | PBANKA_1454100 | 155 S | QKKPSPHFK | 0.99 | unsp |


