

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_1460700 | SP | 0.001357 | 0.998622 | 0.000021 | CS pos: 19-20. VKG-DL. Pr: 0.9297 |
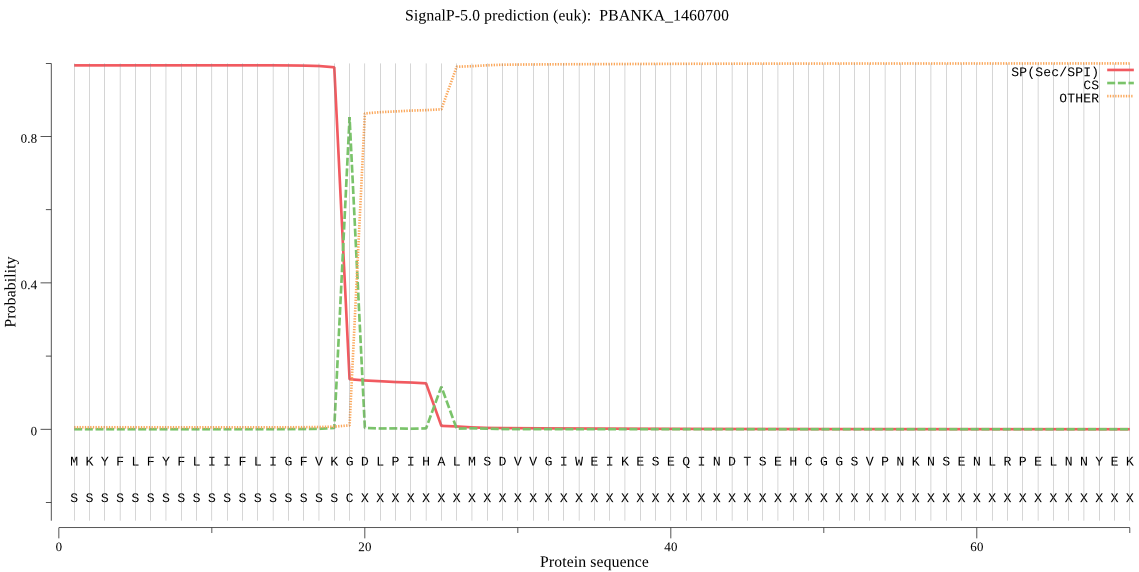
MKYFLFYFLIIFLIGFVKGDLPIHALMSDVVGIWEIKESEQINDTSEHCGGSVPNKNSEN LRPELNNYEKYLQTNYGELKNFKINLTIEKIKLFNDSSSRRIWTYLAVRAVGDNNSVIGN WTMVYDEGFEIRLMGKRYFGFFKYDKKVSEECPSIVENINKMNTECYKTDPTKIRLGWIF YERKQKNHKEKIYQWGCFYAEKITKMPISSFAIHNNIVNNKDHIDTGANFFVVMQRNNYN KIRMNHPELFGSTKISYIDRHKGKENRIYGCKKRESKKMEMDLVLPKEFSWGDPYNNKNF DEIVEDQKECGSCYLISSIYILEKRFEILLSKKYKKNIKMNKLSQECIINLSQYNQACDG GFPFLVGKDIYENGICATMKYGSLDSISSKIETLSRNKKNNNNNNQIYYASDYNYISGCY ECSNEFDMMKEILNNGPIIAAIYATSLLLKLYKLNDHNFIFTNITDENKICDIPNKGFNG WQQTNHAVVIVGWGEHINKNNELIKYWIIRNTWGNQWGYKGYLKYQRGINLNGIESQAVY IDPDFTRGGGKELFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_1460700 | 149 S | DKKVSEECP | 0.992 | unsp | PBANKA_1460700 | 276 S | KKRESKKME | 0.997 | unsp |