

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
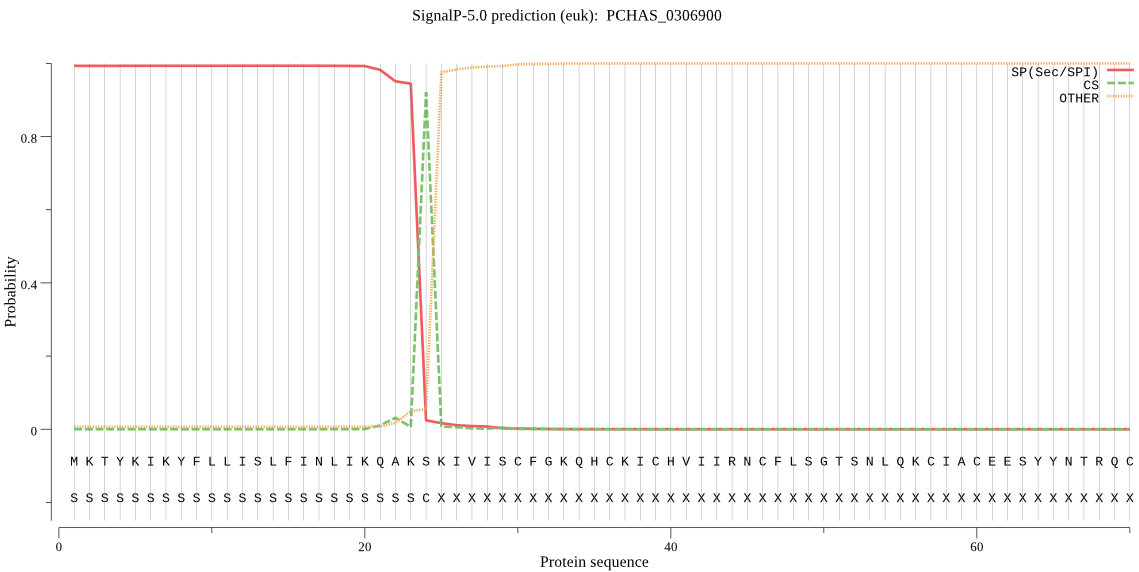
| PCHAS_0306900 | SP | 0.066071 | 0.933839 | 0.000090 | CS pos: 24-25. AKS-KI. Pr: 0.9555 |
MKTYKIKYFLLISLFINLIKQAKSKIVISCFGKQHCKICHVIIRNCFLSGTSNLQKCIAC EESYYNTRQCIRQEGSLFNNKENNALLELKDESLQDSISEDSLDGLVSSILDIAMLRYKN KTTDAGSMDDDYKTKIDNLCLYLNFKDNYENAEKHDQTDAEHVETHIQHIIQMFIHANDN MEHMRNALKNPALCFQNPAEWVQDRLGYKENAEIPSVGVIPEKKLFKPHKHSSLMNSLYN PNSKCNRKYCNRFADPNECEHNIRPLNQGTCGNCWAFASSTTISAFRCRKGLGFAEPSIK YVTLCKNKYLVDEEPPVFGHYNDNICHEGGHLSSYIEILDLSKMLPTSFDVPYNEPIKGG DCPTEVSTWGNLWNGVTALDKILNGYIYKGYFKISFLDYTQAGKTNELINILKDYIIEQG AIFVSMEISKKLSFDHDGEQVMMNCEYGEAPDHALVLIGFGDYIKSSGEKSSYWLIRNSW GSRWGDNGNFKMDMYGPRNCNGRVLFNAFPLLLKMTENNISKPLPNDMSSTDVKIRYNHS DFTKNKKNKTSQRDRNRKVTPNSYDDPYDINPRTVDPSNNNQNDYVNPYYNPDYNPIDNN DRQNNDRHPPFRDSEYDPAYPSMNNNRRFDPNDIINHRNRLRRIFQTDLTVTIGNFVYKR NIYSKRKNEYMEEFSCLRTYSMDTNLDNICRENCEKHIDTCMHSTVIGECLSRNAPNYKC IYCGM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0306900 | 551 S | KNKTSQRDR | 0.994 | unsp | PCHAS_0306900 | 551 S | KNKTSQRDR | 0.994 | unsp | PCHAS_0306900 | 551 S | KNKTSQRDR | 0.994 | unsp | PCHAS_0306900 | 563 S | VTPNSYDDP | 0.995 | unsp | PCHAS_0306900 | 614 S | PFRDSEYDP | 0.998 | unsp | PCHAS_0306900 | 97 S | SLQDSISED | 0.992 | unsp | PCHAS_0306900 | 127 S | TDAGSMDDD | 0.997 | unsp |