

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0307000 | SP | 0.014408 | 0.985506 | 0.000086 | CS pos: 24-25. IKT-QP. Pr: 0.6232 |
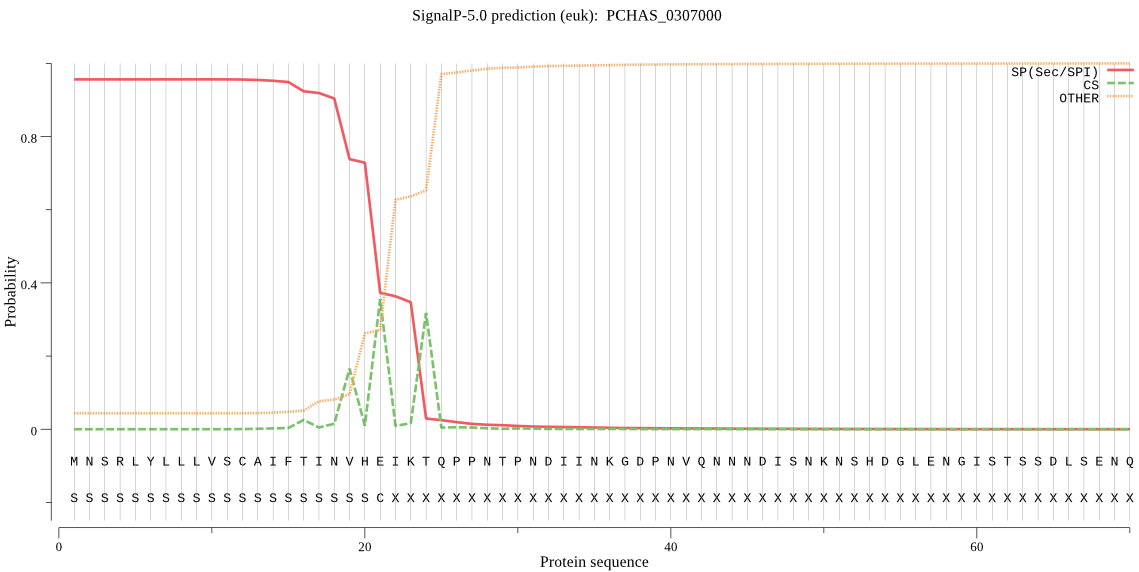
MNSRLYLLLVSCAIFTINVHEIKTQPPNTPNDIINKGDPNVQNNNDISNKNSHDGLENGI STSSDLSENQSNNNTNSESSNSETPDVKTSIKSSLSKDGKGIKVTGTCNEDFRIYFSPYV WIYVTFSEGIIKIESENGVTSSINLNSLNNKCDNKKNDTFKFSAKIKDDILKIKWKTQTN ETDQGHKKDVKQFRLPDLSKEPTSIQIFTANAEEKIIESKTYDIDKNIPEKCSAISANCF LGGSLNIESCYHCTLLAQKYPSDDECFNYISSESKDKINKDTIIKGEDESDENSDEYMLK ESIYKILKKMFTNDSDYCDGSRCNKEMITDIKELDADLRIDLKNYCDILKKVDKSGTLEV HEIASEVEAFNNLVRLLNTHTNENIYILYEKLKNPAICIKNVNEWIIKKRGLVISKEHDV NPYNNPDNPKIKNILNEKYNEENTEAVGNDIIENESDNENEIVNLKNSSNKKLTSAYFNS SRYCNKDYCDRWQDKTSCMSNIEVEEQGECGVCWVFASKLHLETIRCMRGYGHYRSSALY VANCSERDKEEICDVGSNPVEFLQILHDKKHLPLESDYPYSYYRVGGSCPSPKNSWTNLW GNNKLLYYKSRSVDFTSSYGFIALSSSNYQDDLDTYIQIIKNEVRNKGSVIIYIKTNNVI DYDFNGKIIHNLCGDDDDDADHAAAIIGYGNYISEKGQKRSYWLIRNSWGYYWGDEGNFK VDMYGPRHCKYNFIHTVVIFKLDLGEIEAPQKDNQVYNYFAKHIPAFFTNLFYSNYNKRW DEFYAKEELNNYNKDISISGQTDNQVNTNDDVYSEDTNDVSTPSSKNTSIQKKLDVTHVL KYVKNNQTQTSFIKYDDILEIEQEHNCSRVQSIGLKNKNECKSFCLENWSTCKNHPSPGF CLTTLYSAEDCFFCNI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0307000 | 199 S | LPDLSKEPT | 0.992 | unsp | PCHAS_0307000 | 199 S | LPDLSKEPT | 0.992 | unsp | PCHAS_0307000 | 199 S | LPDLSKEPT | 0.992 | unsp | PCHAS_0307000 | 290 S | GEDESDENS | 0.995 | unsp | PCHAS_0307000 | 824 S | VSTPSSKNT | 0.994 | unsp | PCHAS_0307000 | 80 S | NSESSNSET | 0.997 | unsp | PCHAS_0307000 | 90 S | DVKTSIKSS | 0.991 | unsp |