

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0307100 | SP | 0.000472 | 0.999528 | 0.000000 | CS pos: 21-22. VIS-DD. Pr: 0.9792 |
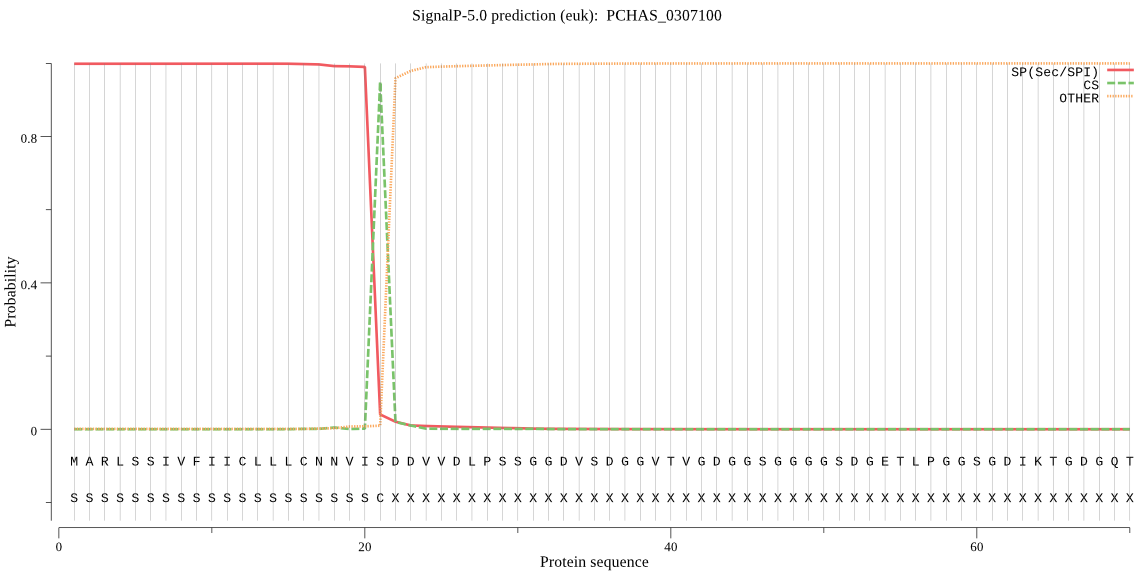
MARLSSIVFIICLLLCNNVISDDVVDLPSSGGDVSDGGVTVGDGGSGGGGSDGETLPGGS GDIKTGDGQTGKGEQGSSNTDQVPGSTDSQDSDQPPKESAPTTPKEPAPTTPKEPAPTTP KEPEPTTPKEPEPTTPKESEPTTPKEPAPTTQKDSGSQSVEKSGITTNDGPAVITGPDSS TGDASLPSAGDQKGEKKENQDPASLPSEDTTDEQTKKETEQESLPTSKGEPESLSEKPSG DPAPKPAEDPASPPAKEPESETPKVSQDATDLNTQKQGEPNKRSRRSPQPQVPETKKVND ITDMESYLMKNYDGVKVIGLCGVYFRVQFSPHLLLYGLTKFSIIQIEPFFERVRIDFEHQ HPIRNKCVPGKAFAFISYVKDNILTLKWKVFVPPADLFANDVVSKQILSSVSEEPSSPQV VDVRKYRLPQLDRPFTSIQVYKANTKEGILETKNYVLKNAIPEKCSKISMDCFLNGNVNI ENCFKCKLLVQNAKPSDECFKYLPSDMKENLNDIKITAQSDEDNKEIDLIESIDILLSKF YKVDLKTKKLGLITIDDFDDVIKVELYNYCKLLKGLDTQKTLENVEMGDEMDVFNNLLRF LKTNEGETKLNLYKKLRNTAMCLKDVNTWAEKKRGLILPEETTEQFASAQSEGFYEEDPD DKVNLLDLFDNDQDEDVVDKDGIIDMSLAIKHAKLKSPYFNSSKYCNYDYCDRWQDKTSC ISNIDVEEQGNCSLCWLFASKLHLETIRCMRGFGHNRGSALYVANCSERKGEQVCNEGSN PLEFLKILEKNRFLPLESNYPYLWKNVSDTCPRPEEHWTNIWGNTKLLYNNMYGQFIKHR GYIVYNSRYFAKNMDVFIDIVKREIRNKGSVIAYIKTKDVIDYDFNGRYISNICGDSHPD HAVNIIGYGNYISDKGEKRTYWLIRNSWGYYWGHEGNFKVDVLGPDDCVHNFIHTAIVFK IDMEPDNHGGIKNKDQSIDENKKSYFPQLSSNFYHSLYYNNYEGHEAKSDDENDRDYDNA DVSGESETSDDSSGDSSENGSEESQPQSGGSENPVSSPEAATSIPAPVDSPTPNIATIEK TIQILHILKHIENYKMTRGLVKYDNLNDTKHDYACARSSSNDPKKVNECKQFCEENWERC KKHYSPGYCLTALAGKSSCLFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0307100 | 119 T | PAPTTPKEP | 0.996 | unsp | PCHAS_0307100 | 119 T | PAPTTPKEP | 0.996 | unsp | PCHAS_0307100 | 119 T | PAPTTPKEP | 0.996 | unsp | PCHAS_0307100 | 127 T | PEPTTPKEP | 0.995 | unsp | PCHAS_0307100 | 135 T | PEPTTPKES | 0.995 | unsp | PCHAS_0307100 | 143 T | SEPTTPKEP | 0.995 | unsp | PCHAS_0307100 | 180 S | GPDSSTGDA | 0.996 | unsp | PCHAS_0307100 | 188 S | ASLPSAGDQ | 0.993 | unsp | PCHAS_0307100 | 215 T | TDEQTKKET | 0.992 | unsp | PCHAS_0307100 | 227 S | SLPTSKGEP | 0.996 | unsp | PCHAS_0307100 | 287 S | RSRRSPQPQ | 0.997 | unsp | PCHAS_0307100 | 410 S | QILSSVSEE | 0.993 | unsp | PCHAS_0307100 | 412 S | LSSVSEEPS | 0.994 | unsp | PCHAS_0307100 | 520 S | ITAQSDEDN | 0.997 | unsp | PCHAS_0307100 | 977 S | NKDQSIDEN | 0.996 | unsp | PCHAS_0307100 | 1009 S | HEAKSDDEN | 0.996 | unsp | PCHAS_0307100 | 1032 S | TSDDSSGDS | 0.997 | unsp | PCHAS_0307100 | 1033 S | SDDSSGDSS | 0.994 | unsp | PCHAS_0307100 | 1036 S | SSGDSSENG | 0.996 | unsp | PCHAS_0307100 | 1041 S | SENGSEESQ | 0.994 | unsp | PCHAS_0307100 | 1057 S | NPVSSPEAA | 0.995 | unsp | PCHAS_0307100 | 1119 S | CARSSSNDP | 0.996 | unsp | PCHAS_0307100 | 103 T | SAPTTPKEP | 0.995 | unsp | PCHAS_0307100 | 111 T | PAPTTPKEP | 0.996 | unsp |