

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0307200 | SP | 0.005311 | 0.994689 | 0.000000 | CS pos: 22-23. INC-RP. Pr: 0.9115 |
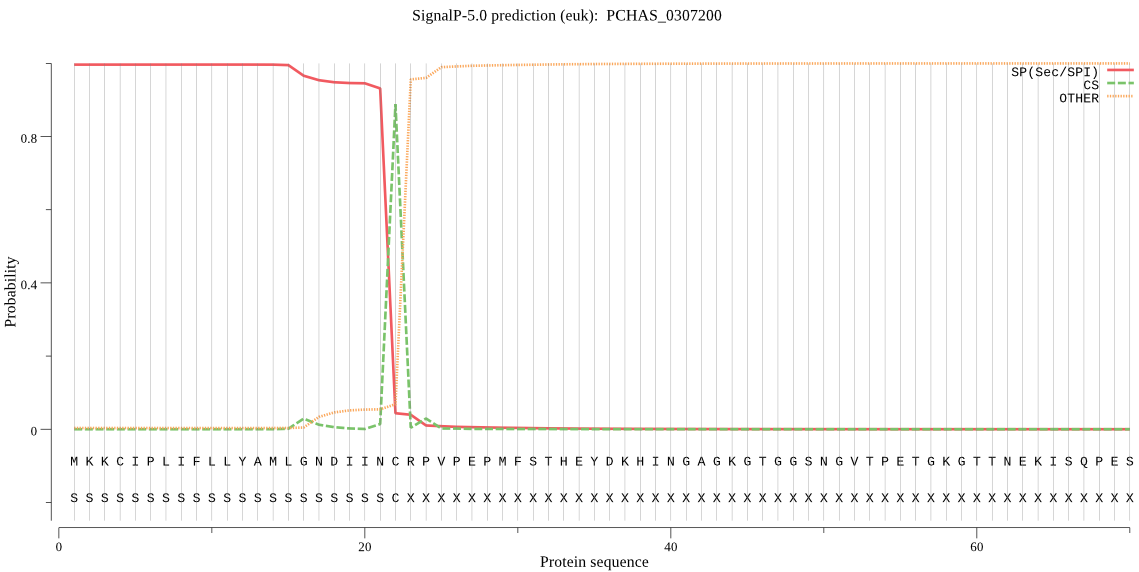
MKKCIPLIFLLYAMLGNDIINCRPVPEPMFSTHEYDKHINGAGKGTGGSNGVTPETGKGT TNEKISQPESEKDKAPESSPDSSTLPSPGSGPSPSSPESSDSSNSSDSSDPSASSDKSKN PSGTGTDSSSSSQGQDAKNQTQPQPQPQPQPQPQPQTDDNPSSKTNTDNVDASQTLSVTE PAANSTQVVAPKNVQTGSVESALLKNFNGVKVTGACGDEAAIFLEPYIYINIDSKNTNIN LSANFSNLFGQHITVFAGQTPMGNVCHEEDDSLRYKMLVYLQDDILTIKWSVFPSNPESN SCKYCTGHTNENEHVDVKKFRLPRLPTPLTTIQVHSLLNKEDKVIIRSKDYALKNEMPKK CDQVASKCFLNGQTDIEQCYTCSLLTEKIPTTDECYNYSSPLVREYTQVLTTAQSDEDNV EKNNTALVESINNLLNGVYKVDEHNNKVLIDEEELSNDLKKELTYYCQVLSEIDTSGTLE NYKVGNSVEIFNNLAKLLKNHENENKLYLINKLKNPAICMKDVEQWVVNRKGLKLPVALD DAEKKTDNAIKFEEGPDGIVDLTKTVDGNPITTTAVLANKLDAFCNNEFCDRLNDKTSCI SNINVEEQGNCATSWVFASKLHLESSICMKGYDHTSASALYVANCSKKDSKDKCLAGSNP LEFIDILQTNNYLPTESNYPYNYKNVGDACPETNESWVNLFDKVKLEDSNKDNKSIIKGY KSYESKNYKTNMNEFVDIVKNNIKKLGSVIAYVKFNDSMGYDFNGSKVHKLCGSATPDMA VNIIGYGKYINENNEVKSYWLVRNSWGKHWADEGNFKVDIETPEDCEDNFIHTALVFDLD LPVVAKNTNEAPIYNYYLKTSPSFYKNLYYNHVNKTHGEVSDDAAIYGQAEPEAPGKNNQ SGSTTDTTKVTTANSESGSDAGGKTKTGESGTSATGKETPAVAGTDGQTNKVTDSSAGQS PSVPTGASGPTPQAPAGPQSPSNPASGVTQDPSNPTPGSNGEQADQAVTETEADATQNTE VFHVLKVVRNKKVQTSLLKYGTYTEMGQHSCARAMSTDPTKKEECVAFCNNNWDDCYFDL YPGYCLNKLKGDNVCNFCAV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0307200 | 78 S | KAPESSPDS | 0.995 | unsp | PCHAS_0307200 | 78 S | KAPESSPDS | 0.995 | unsp | PCHAS_0307200 | 78 S | KAPESSPDS | 0.995 | unsp | PCHAS_0307200 | 96 S | PSPSSPESS | 0.998 | unsp | PCHAS_0307200 | 122 S | SKNPSGTGT | 0.994 | unsp | PCHAS_0307200 | 131 S | DSSSSSQGQ | 0.99 | unsp | PCHAS_0307200 | 163 S | DNPSSKTNT | 0.991 | unsp | PCHAS_0307200 | 415 S | TTAQSDEDN | 0.996 | unsp | PCHAS_0307200 | 646 S | VANCSKKDS | 0.995 | unsp | PCHAS_0307200 | 709 S | KLEDSNKDN | 0.997 | unsp | PCHAS_0307200 | 903 S | NQSGSTTDT | 0.993 | unsp | PCHAS_0307200 | 917 S | ANSESGSDA | 0.995 | unsp | PCHAS_0307200 | 66 S | NEKISQPES | 0.996 | unsp | PCHAS_0307200 | 70 S | SQPESEKDK | 0.998 | unsp |