

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
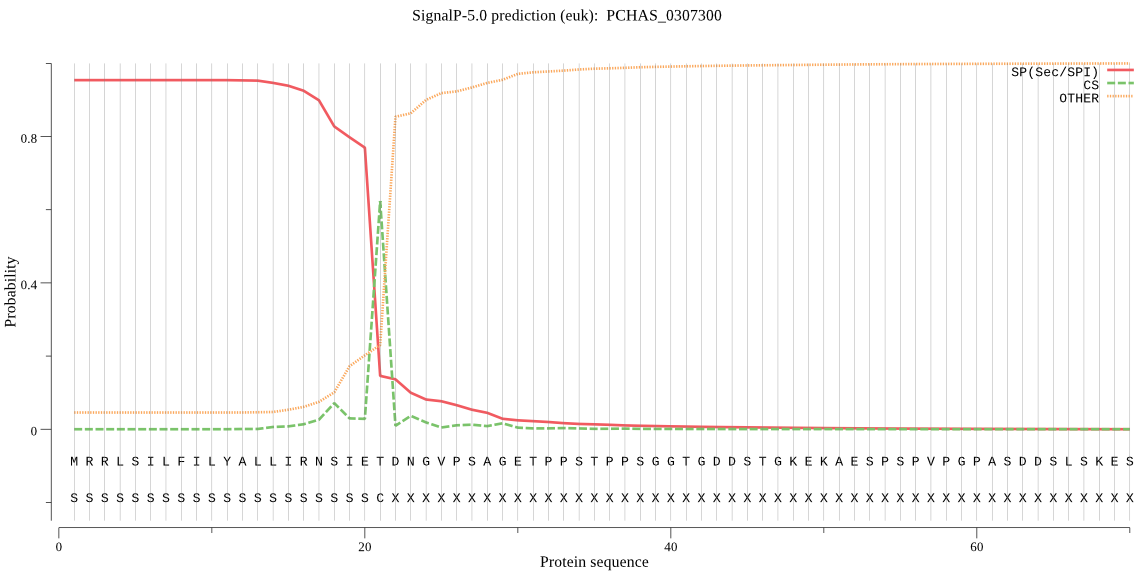
| PCHAS_0307300 | SP | 0.007536 | 0.992449 | 0.000015 | CS pos: 21-22. IET-DN. Pr: 0.7570 |
MRRLSILFILYALLIRNSIETDNGVPSAGETPPSTPPSGGTGDDSTGKEKAESPSPVPGP ASDDSLSKESVNTGGSELKTPSSSDGGPKATEEDRKEAGETEKKVSESDKPKEGEPASDK TQQDSTKQDASPSSVDGQNPQAKGETGSPGPVQPPPESGELPKNDSEQKISEPGVVGSDV SQPEAVKSDASTLPGSVSDNSSKGGDASDPQKSKDAKKVDNESSGKSDDPNKNTKQGDSI SEPDSGLKTDTDGTENPQEGQAKKGVEGTNQEENPPKAGSEPTPGKAEVTKDPPTPANKV PGSEETETLTSGPTLDPQSQNLPSSTDNPLPTGQESGNTRDIAQGDDPSVDSTPDATPAV FLENFNVKSAMLKNYKGLKITGECNADLSIFFVPYILIDVNTKTNQIFIMSTKKKVMGTT KGVATPSIDFKSEGDTLQNVCGENKTFKIVVYFDGDVLYVKWKVYDPSVQTEPNTNVDVR KYRLKNFETPITSIQVHSIKGGSKTVTFETKNYTINDHLPEKCDEIASDCFLHGHAEIEK CYKCALLYQKTPLTDECYNYVPRDYKESLKADLARAQSDDEINYEVMEHIENILDGIYNV NDNNKELKTFEELSDETKKDILLYCKELKESDVTGTLEDYASGDAEDVYANLTKLITNNM EDSISKLKSKLMNPAICLKDVNQWGEKKTGLVLPELSSNNVDNSNENDDTKTDLESDEKL KDEGFDGVIDLPLPHENEFAGYTTIEDFQYCNNEYCDRLKDKNSCISKLNVEEQGNCATS WLFASKLHLDTIGCMKGHENFSASALYVANCSKTDSKDKCLTGSNPLEFLNIIDENKFLP TTSNLPYSYKKVGEECPQTMENWTNLWKDVKLLKSENNDKSLNANGYISYQSEHFKENFN EYVNLIKQEIQNKGSVIAYVNSKDINSYDFNGSKVHKLCGSATPDLAVNIIGYGNYANEN NEVKSYWLVRNSWGKHWADEGNFKVDIETPENCEHNFIHTFATFNIYVPFVKKNTQTESE INLYYSKNSPDFYNNLYLKNLDADNTNEDSLIQGQDEPAKVPTSGDTEQSTKDVDPKSSQ KAGTEEKLGSAELKETIQDSGSSSKATLQSTDLSTNPPPKGQEQVQEQVQEQEQKAKQED KGKTDKKAGEGTDSKAGQAGKKDGTTEDEASKTKKDSPPAKPEGPQASQPQADQPQVVQP QAPGQPASGAKARLPAGPKLEFMHVLKQIKNGKVKMNLVKYGNELNLIDERACSRVYAVN PEKKSECEEFCVNKWNECKDSEVPGYCLADLYNSNNCYFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0307300 | 106 S | EKKVSESDK | 0.998 | unsp | PCHAS_0307300 | 106 S | EKKVSESDK | 0.998 | unsp | PCHAS_0307300 | 106 S | EKKVSESDK | 0.998 | unsp | PCHAS_0307300 | 108 S | KVSESDKPK | 0.995 | unsp | PCHAS_0307300 | 125 S | TQQDSTKQD | 0.993 | unsp | PCHAS_0307300 | 131 S | KQDASPSSV | 0.992 | unsp | PCHAS_0307300 | 196 S | TLPGSVSDN | 0.996 | unsp | PCHAS_0307300 | 201 S | VSDNSSKGG | 0.996 | unsp | PCHAS_0307300 | 239 S | KQGDSISEP | 0.997 | unsp | PCHAS_0307300 | 245 S | SEPDSGLKT | 0.992 | unsp | PCHAS_0307300 | 324 S | QNLPSSTDN | 0.991 | unsp | PCHAS_0307300 | 352 S | PSVDSTPDA | 0.993 | unsp | PCHAS_0307300 | 432 S | IDFKSEGDT | 0.995 | unsp | PCHAS_0307300 | 1070 S | DTEQSTKDV | 0.996 | unsp | PCHAS_0307300 | 1177 S | TKKDSPPAK | 0.991 | unsp | PCHAS_0307300 | 27 S | NGVPSAGET | 0.994 | unsp | PCHAS_0307300 | 53 S | EKAESPSPV | 0.992 | unsp |