

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0406600 | SP | 0.072692 | 0.926487 | 0.000821 | CS pos: 20-21. IET-KQ. Pr: 0.7410 |
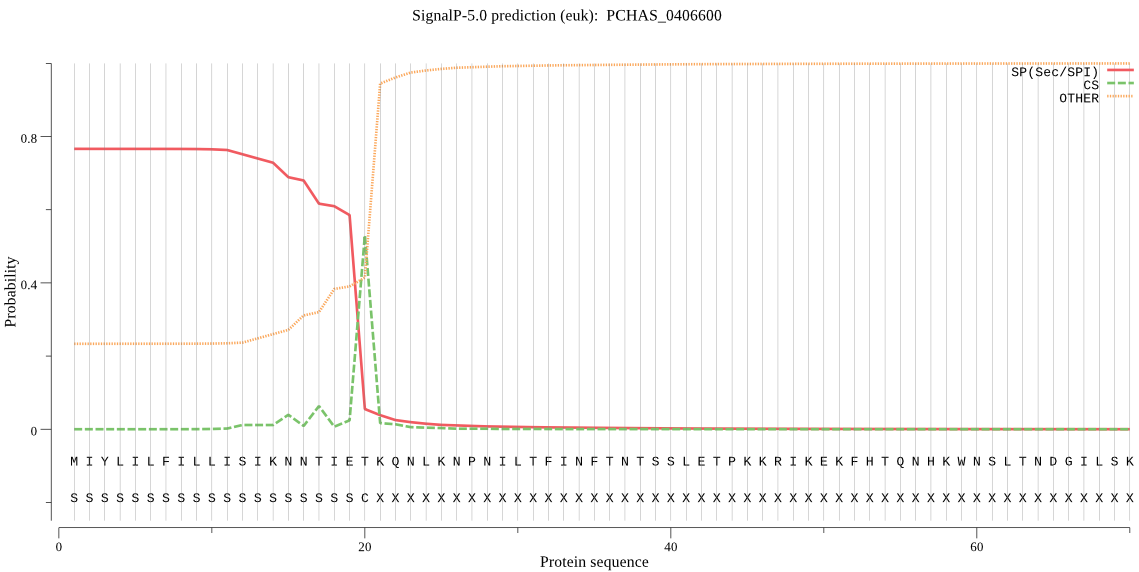
MIYLILFILLISIKNNTIETKQNLKNPNILTFINFTNTSSLETPKKRIKEKFHTQNHKWN SLTNDGILSKNIEGLNNGKNISKNLNETTTNTNNDVTKVDQGAESKIRQNVMEKMLEKIM KKKKYKKLEKLLCCNNSIKAMKKNIKLYFFKKRIIYLTDEINKKTSDELIKQLLYLDNIN HNDIKIYINSPGGSINEGLAILDIFNYIKSDIQTISFGLVASMASVILASGKKGKRKSFP NCRIMIHQPLGNAFGHPHDIEIQTKEILYLKKLLYYYLSKFTNKSEETIEKHSDRDYYMN ALEAKEYGIVDEVVETKLPHPYFPDLTKS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0406600 | 166 S | NKKTSDELI | 0.994 | unsp | PCHAS_0406600 | 293 S | IEKHSDRDY | 0.998 | unsp |