

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0412900 | SP | 0.037577 | 0.959770 | 0.002653 | CS pos: 23-24. CDA-KV. Pr: 0.8801 |
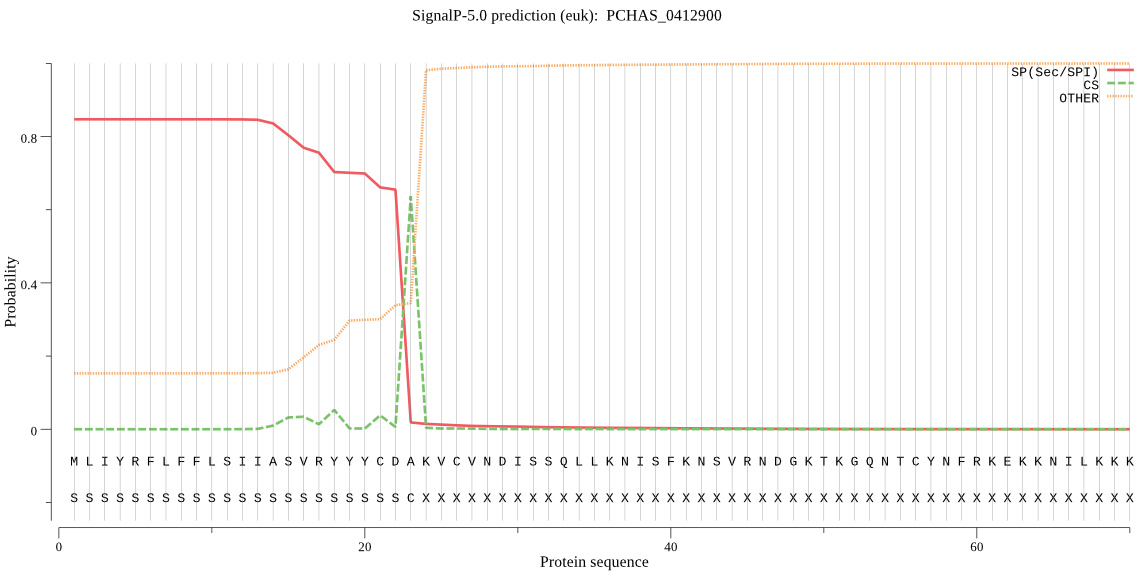
MLIYRFLFFLSIIASVRYYYCDAKVCVNDISSQLLKNISFKNSVRNDGKTKGQNTCYNFR KEKKNILKKKIKKKYSLNNSKGYTQKKKKYNLYFLNSSKFTDIKENVKAWFNLDYIENHY AYSYIKSFARIPPITKIYLINTFILSVLIHLNKNVYKYILYDFDKIFKNGEIWRLVTPYF YIGNLYLQYILMFNYLNIYMSSVEIAHYKNPEDFLIFLTYGYISNIFFTIIASMYNENVM DLKKHIRKIRSLITAMPSTNNKDDNKFVQKETYNHLGYVFSTYILYYWSRINEGSLINCF DLFFIKAEYIPFFFVIQNILLYNEFSFFEVASILSSYTFFTYEKYLKSNFLRALFRGFLK FIRVYPMYNMYKEEYE