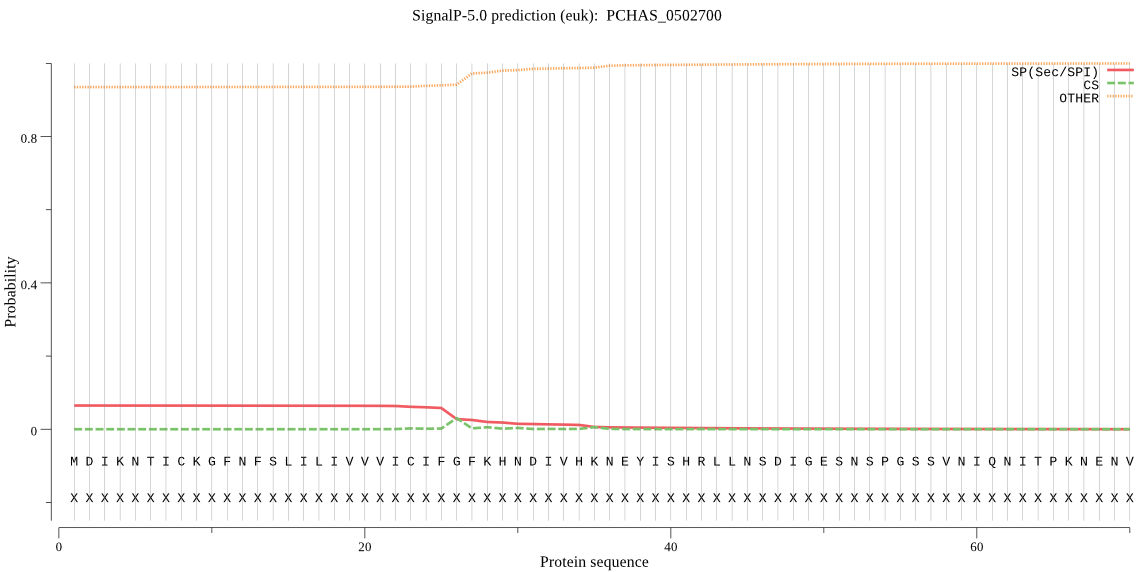
Fasta :-
MDIKNTICKGFNFSLILIVVVICIFGFKHNDIVHKNEYISHRLLNSDIGESNSPGSSVNI
QNITPKNENVDTTNSNHEVDDEMKPMGNNAESDKGNDAEQYENKKDMHINNDNKSNTLES
SDEKNGNKEINMSVSSDTIENVEEKSERNKSNYKTVNEFTGLTLDLYFTEHNNIYTNVKV
GNQLLKLSLNSRLEGMYVFMDDSISCYQKDENSKRKCYDPSLSETALWCSNNMICLPAIL
SMPYECYADKRLNINNKVEYPNIYYDALKYSASHIEGFDTIELMDLKHVNKKVEGKNDNG
EVEPNNLNTFETADIKLIVDLSIYNNWDLFKDTDGIMGLAGNELSCRNKSVWNEILEKNN
SLFGIDINLPENATKKYINNSSEKNGRNINFYNALVENNKKNKNMNNTGFVTNIQESVIK
LFSSKTKNGNPNMLNGEMLPSEIHIGDYKKEYEPILWSEPRERGGIFSDSFMQFTIYNLE
VCNNNIFGKNSSNWQGAIDLSSKCLVLPKMFWLSLMQYLPVNKNDERCIPSNKEIEFNED
TVPRMCSIDDKYRPLPVLKFSFSDNDIVSNNNINNNDNGHGENIKNIHIPLDNLIIKEDG
PNNNYLCIIPDVREGTSNGNSGRTTKPLIKFGTYVLNNFYVVVDQENYRVGLSNKKSFHH
SNDKCTQKVECIGDQIYEPALNICIDPDCSIWYFYTLNHETKTCESVSSRFYVFIMVLFI
LLIVDIQSYYLYRRSVRTAKVSSR