

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0601700 | OTHER | 0.999853 | 0.000124 | 0.000023 |
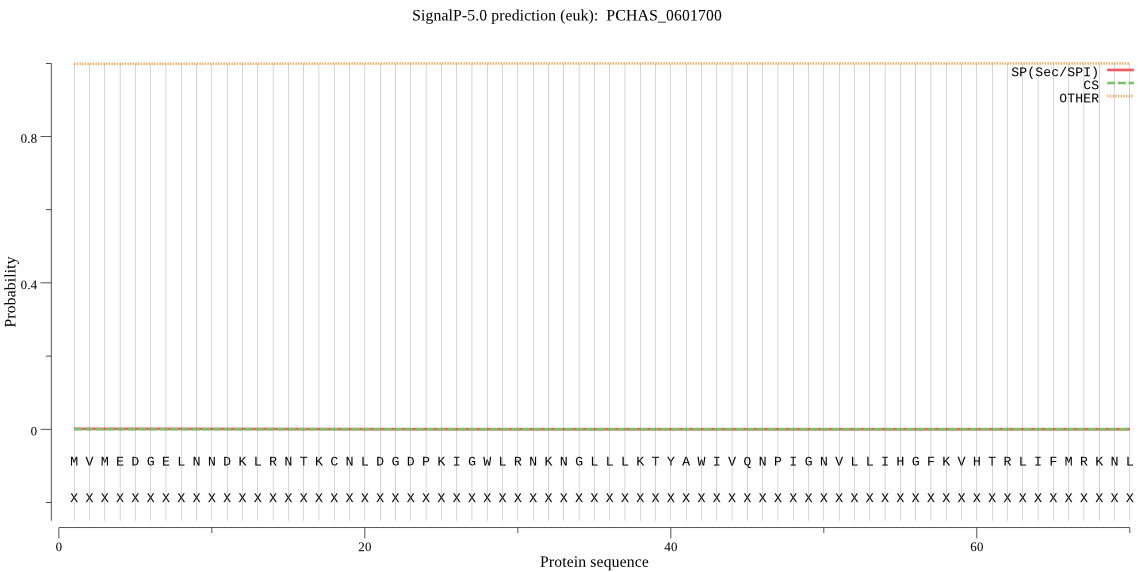
MVMEDGELNNDKLRNTKCNLDGDPKIGWLRNKNGLLLKTYAWIVQNPIGNVLLIHGFKVH TRLIFMRKNLKMSNNNEGLVIDNNNRYIYKDSWIEKFNQNGYSVYALDLQGHGESQSFGK IRGDFSCFDDLVDDVFQYMNQIQGETPSDNKKGDEPHNIVTTKKKKLPMYIVGYSIGGNI ALRILMLLEKEKEDRTKARNSNNYKNSNTRLSNSTNINDIDNGMYDMNNSNNYGFDKSCS SISAATNAIASDNHEECYDSFDKFNIKGCVSLSGMIRIKSVLDPGNISFKYFYLPLADFL AYVLPNPRILSESRYKKPGHFANMCKHYIFRNINGKKYKTMSEFIKATVKMDRNINYIPK DIPLLFVHSKDDSICSYEWTVSFYNKLNVNDKELHVVDGMNHDTTIKPGNEEILKKIIDW ICNLRTNDENK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_0601700 | 280 S | IRIKSVLDP | 0.993 | unsp |