

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0613700 | SP | 0.032931 | 0.965355 | 0.001714 | CS pos: 17-18. CVC-KK. Pr: 0.8994 |
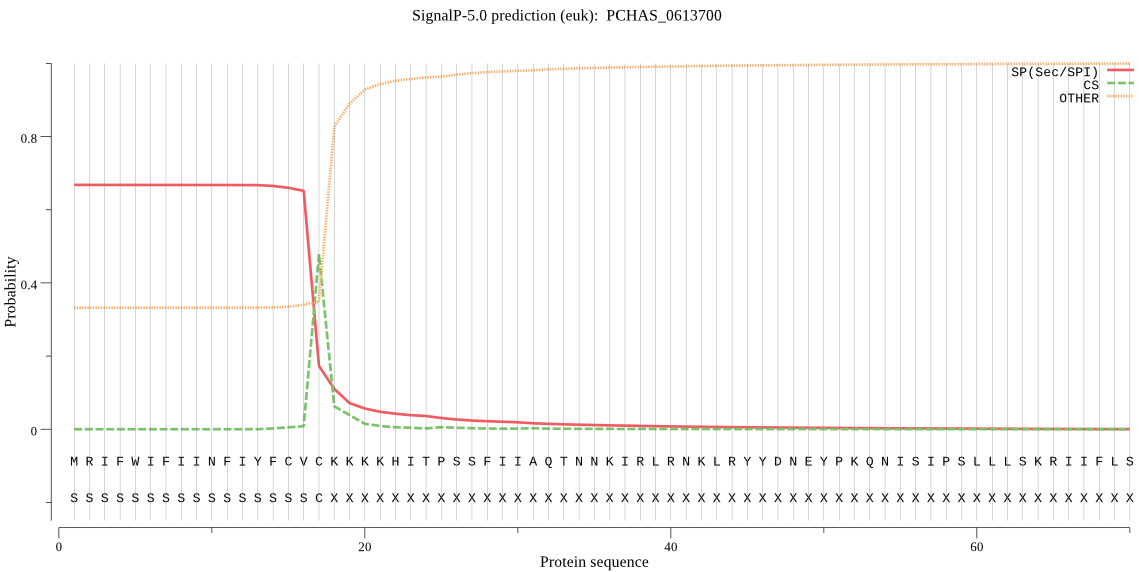
MRIFWIFIINFIYFCVCKKKKHITPSSFIIAQTNNKIRLRNKLRYYDNEYPKQNISIPSL LLSKRIIFLSSPIYPHISEQIISQLLYLEYESKRKPIHLYINSTGDLENNKIVNLNGITD VISIVDVINYISSDVYTYCLGKAYGIACILASSGKKGYRFSLKNSSFCLKQSYSIIPFNQ ATNIEIQNKEIMNTKKKVIDIIAKNTEKDSNIISEILDRDRYFNAKEASDFNLIDHILEK Q