

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
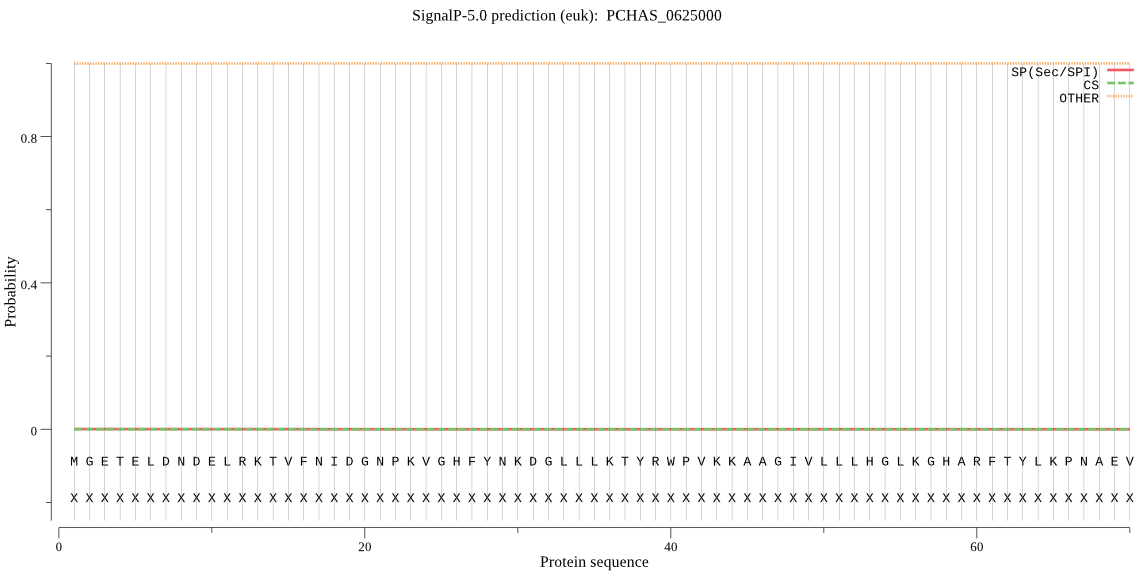
| PCHAS_0625000 | OTHER | 0.999884 | 0.000057 | 0.000059 |
MGETELDNDELRKTVFNIDGNPKVGHFYNKDGLLLKTYRWPVKKAAGIVLLLHGLKGHAR FTYLKPNAEVIDNNEVLVIDDDNYYVYSGSWVEKFNQNEYSVYAMDLQGHGESEARKNLR GHFKRFNDLVDDVLQYMNQIQDEIANDNKIDDESYSIVPAKKQKLPMYIIGYSMGGNIAL RILQILNKAKVKNNSKLEDTVIYAKADTLTGDCTNIYDSDNDNDNDNDNDDDDDDDDDDV YYDVYDGDNVNDNNPTNKATNVSEVVDINNVSNDNTTKHYACTYCIANGSTCDNDSVSTT TGGSDIDNFSVSADDSVAASTSTEDNVVASTSTEYNEVPSTSTEDNVVASTSTEDNVVVS TSTEDNEVPSTSTEDNVVASTSTEDNVVVSTSTEDNEVPSTSTEDNEVVSTSTEDNEVPS TSTEDNEVASTSTEDNEVPSTSTEDNVVASTSTEDNVVVSTSTEDNEVPSTSTEDNVVAS TSTEDNVVVSTSTEDTVVPSTSTEDNAVASTSTDDNAIASTSTDDNLIASTSTEDNAVAS TSTEDNAVASTSTEDNVVASTSTDDNAVASTSTEDNAVASTSTDDNAVASTSTEDIVVVG TSTDDNDKNDDKYNCLDKLNIKGCVSLAGMVSFERIAQPGTYLFNYVYLPVTHFLSYIAP NLETKSELPYKGYPFIDNLCKLDKYRGNRGVTVKCVYELIKAMGALNDDINYIPKDIPIL FVHAKGDSICYYKGVEMFYDRLHSDNKELFPVDNMDHSITLEPGNEQVLEKIIKWLNNLK NNNKNAIENQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0625000 | 312 S | NFSVSADDS | 0.995 | unsp | PCHAS_0625000 | 312 S | NFSVSADDS | 0.995 | unsp | PCHAS_0625000 | 312 S | NFSVSADDS | 0.995 | unsp | PCHAS_0625000 | 322 S | AASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 342 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 352 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 362 S | VVSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 372 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 382 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 392 S | VVSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 402 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 412 S | VVSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 422 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 432 S | VASTSTEDN | 0.998 | unsp | PCHAS_0625000 | 442 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 452 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 462 S | VVSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 472 S | VPSTSTEDN | 0.998 | unsp | PCHAS_0625000 | 482 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 492 S | VVSTSTEDT | 0.998 | unsp | PCHAS_0625000 | 502 S | VPSTSTEDN | 0.997 | unsp | PCHAS_0625000 | 512 S | VASTSTDDN | 0.994 | unsp | PCHAS_0625000 | 522 S | IASTSTDDN | 0.991 | unsp | PCHAS_0625000 | 532 S | IASTSTEDN | 0.996 | unsp | PCHAS_0625000 | 542 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 552 S | VASTSTEDN | 0.998 | unsp | PCHAS_0625000 | 572 S | VASTSTEDN | 0.997 | unsp | PCHAS_0625000 | 582 S | VASTSTDDN | 0.994 | unsp | PCHAS_0625000 | 592 S | VASTSTEDI | 0.997 | unsp | PCHAS_0625000 | 602 S | VVGTSTDDN | 0.994 | unsp | PCHAS_0625000 | 219 S | NIYDSDNDN | 0.997 | unsp | PCHAS_0625000 | 304 S | TTGGSDIDN | 0.995 | unsp |