

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
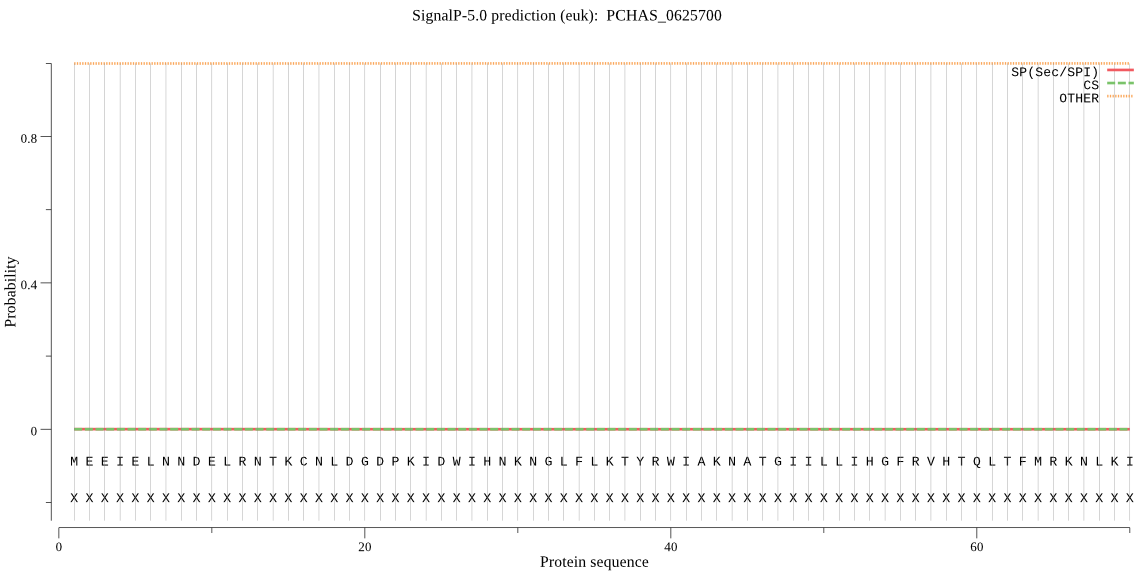
| PCHAS_0625700 | OTHER | 0.999939 | 0.000017 | 0.000044 |
MEEIELNNDELRNTKCNLDGDPKIDWIHNKNGLFLKTYRWIAKNATGIILLIHGFRVHTQ LTFMRKNLKIANGNEELVVDNNNCYIYKDSWIENFNKNGYSVYALDLQGHGESQTWKNIR DNFSSFDDLVDDVIQYMHHIQDEISNDNQTDDESHDIVTTKKKKLPMYIIGHSMGGNIAL RILQLLNKEKGNDIDTGESNNYKNIYTMLNNSTNINEIGNDMNNSKDYDSDNSCASTSDT TNAIASDKHEGCYNCLDKFNIKGCVSLSGMIRIKSKRDPGNKSYMYFYLPIIKFLSCFAP RARISLESRYNMFGYYGNKFRLYKILNINQTKFKCMSELIKATITLDRNIKYMPKDVPLL FVHSQDDSICSYERAVAFYNKVNVSKKELYPVDGMDHGTPTRPGNEEILKKIIDWISDLR SKDEDE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0625700 | 385 S | KVNVSKKEL | 0.994 | unsp | PCHAS_0625700 | 385 S | KVNVSKKEL | 0.994 | unsp | PCHAS_0625700 | 385 S | KVNVSKKEL | 0.994 | unsp | PCHAS_0625700 | 421 S | SDLRSKDED | 0.995 | unsp | PCHAS_0625700 | 236 S | NSCASTSDT | 0.996 | unsp | PCHAS_0625700 | 275 S | IRIKSKRDP | 0.997 | unsp |