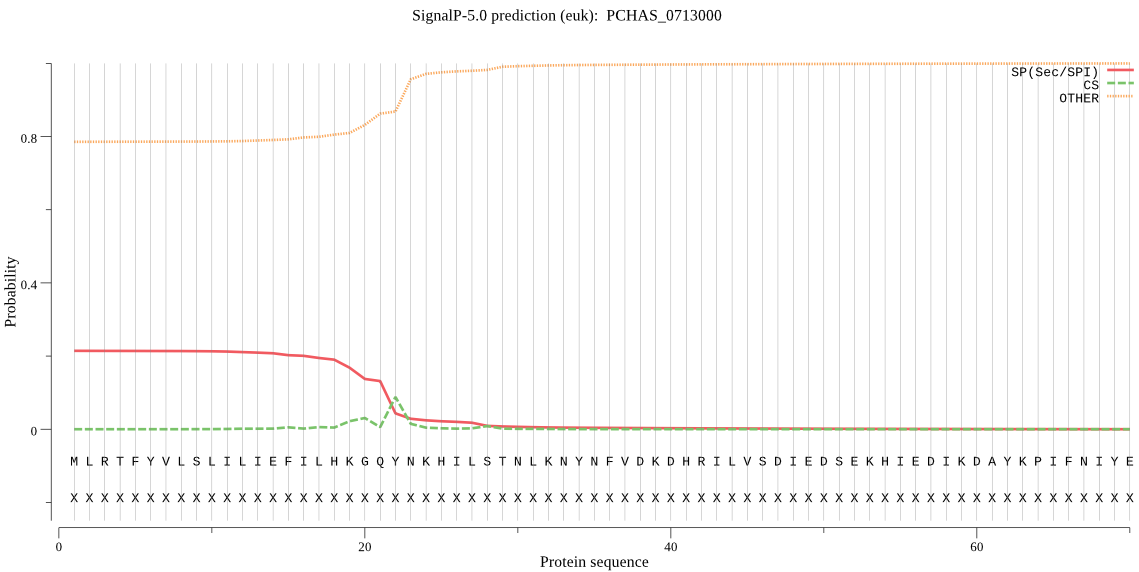
Fasta :-
MLRTFYVLSLILIEFILHKGQYNKHILSTNLKNYNFVDKDHRILVSDIEDSEKHIEDIKD
AYKPIFNIYEISAEFHKKKYIANKKKKQKDGSEQSIEKKINSEGDERSQLNQNEGITFLE
LSDRHPDAENDDHDDLIEERDDTLEQEDSAENNDGILPRGKIYEQDEENKNINTIPSDEN
SNEKVYDKKSDISLKDKIDYKERGYGNFKSKEKNLDENIKTYTFDHYKIITNSENILNDI
KVDASDISKLSINSLNIEYNEKNKAEYTHQRHIILSNEGNRRYKIFLMTKNPKFTKTDDI
PQPEMNFIQTKAVQNEGATEEEEKYYNENLYSGFGTLDFEKGYSKTKKKIDNGHASGVND
QTSNHQNIEKNDSHENEKHNHGFIGKVRSFFSFLSVSNSQKDDNIESGKKNEESNNADSK
TKLNEKTDDTTRKDSLNEFLSVDKLTDQYLLNLKNKNVKEQELILIFQGDLDLHSKEMRK
VIDEANAKLTNYLNMHFKDIKNMNYDISSPINFVCFFIPTVFDISNLNILKEALIMLHNE
LKDYIDNWNFSNTYIAFDRNYEDEDIKKEDIDNVMNKLNEHGEKYIKKPKKLYNIKYSFL
RKIWGLESIISLYKGKNKKEADIEERILNALPKELKEYSTWNLSFIRVFNAWLLSGYGNK
NVKICVIDSGIDKNHIDLASNVYIPKYSDKYEMTDEFFDFMVQNPSDTSGHGTHVSGIAG
ASANSLGMVGVAPNIKLISLRFIDGDNYGGSFHVIKAINVCILNKSPIINASWGSRNYDT
NMYLAIQRLKYTFKGKGTVVVAAAGNENKNNDLYPIYPASYKLQNVYSVGAINKFLQISP
FSNYGAKSVHILAPGHHIYSTTPMNTYKTNTGTSMAAPHVSGVAALIYSVCHNQGFIPEA
EEVLEIITRTSIKIVTREKKTMHNSLVNAEAAVLTTLLGGLWMQMDCHFAKFYLNKDKQK
SIPVVFSAYKDGMYETDIIIGIKPVDTNSKEYGEIVIPIKILTNPNLKDFSASPRIGKKI
HIDANESIDDDILSYICENALYNLYEHDNTFLISSLILFFLGIILIVIVSIVFFFKHHQN
KHRDYEQYMNQKMADRAYDIKYNFNDVGPGGIKKSCSLDGNINNHRDTQRFTIVQNEDNM
YVLKKKSSIKSKYEARNELTKRPLIKRPIVKHKDTNVNFNNMEALYESHGNLSE