

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
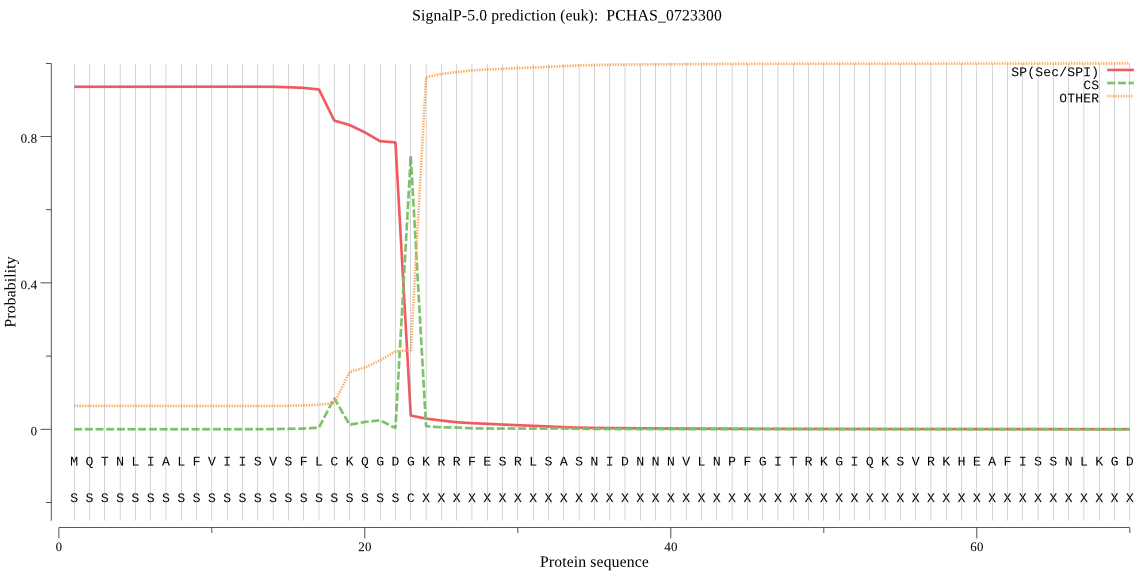
| PCHAS_0723300 | SP | 0.007540 | 0.991754 | 0.000705 | CS pos: 23-24. GDG-KR. Pr: 0.9439 |
MQTNLIALFVIISVSFLCKQGDGKRRFESRLSASNIDNNNVLNPFGITRKGIQKSVRKHE AFISSNLKGDDILKRDTKKGYKSKINENKDENKKYDQGRIQNRLNYSTNDQDNINNYFFS KNENKSNNGRNKANQLFMSDEEYTINSDDYTEKAWEAISSLNKIGEKYESAYVEAEMLLL ALLNDSPDGLAQRILKESGIDTDLLSHDIDLYLKKQPKMPSGFGEQKILGRTLQTVLSTS KRLKKEFNDEYISIEHLLLSIISEDSKFTRPWLLKYNVNYEKVKKAVEKIRGKKKVTSKT PEMTYQALEKYSRDLTALARAGKLDPVIGRDEEIRRAIQILSRRTKNNPILLGDPGVGKT AIVEGLAIKIVQGDVPDSLKGRKLVSLDMSSLIAGAKYRGDFEERLKSILKEIQDSEGQV VMFIDEIHTVVGAGAVAEGALDAGNILKPLLARGELRCIGATTVSEYRQFIEKDKALERR FQQILVDQPSVDETISILRGLKERYEVHHGVRILDSALVQAAILSDRYISYRFLPDKAID LIDEAASNLKIQLSSKPIQLDNIEKQLIQLEMEKISILGDKQSASLINKSSSGNDDNNIS TDYTQSQNFIKKRINEKEINRLKTIDHIMNELRKEQKNILESWTSEKMYVDNIRAIKERI DVVKIEIEKAERYFDLNRAAELRFETLPDLEKQLKTAEENYVNDIPERNRMLKDEVTSED IMNIVSISTGIRLNKLLKSEKEKILNLENELHKQIIGQDDAVKIVARAVQRSRVGMNNPK RPIASLMFLGPTGVGKTELSKVLADVLFDTPDAVIHFDMSEYMEKHSISKLIGAAPGYVG YEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVIDEGKLSDTKGNVANFRNTIII FTSNLGSQSILDLANDPNKKEKIKEQVMKSVRETFRPEFYNRIDDHVIFDSLTKKELKEI ANIEITKVANRLFDKNFKISIDDAVFSYIVDKAYDPSFGARPLKRVIQSEIETEIAIRIL NETFVENDTIRVSLKDQKLHFSKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0723300 | 378 S | DVPDSLKGR | 0.991 | unsp | PCHAS_0723300 | 378 S | DVPDSLKGR | 0.991 | unsp | PCHAS_0723300 | 378 S | DVPDSLKGR | 0.991 | unsp | PCHAS_0723300 | 530 S | DRYISYRFL | 0.993 | unsp | PCHAS_0723300 | 592 S | NKSSSGNDD | 0.994 | unsp | PCHAS_0723300 | 739 S | KLLKSEKEK | 0.993 | unsp | PCHAS_0723300 | 930 S | QVMKSVRET | 0.997 | unsp | PCHAS_0723300 | 980 S | NFKISIDDA | 0.993 | unsp | PCHAS_0723300 | 1009 S | RVIQSEIET | 0.991 | unsp | PCHAS_0723300 | 1033 S | TIRVSLKDQ | 0.998 | unsp | PCHAS_0723300 | 32 S | ESRLSASNI | 0.996 | unsp | PCHAS_0723300 | 198 S | ILKESGIDT | 0.997 | unsp |