

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
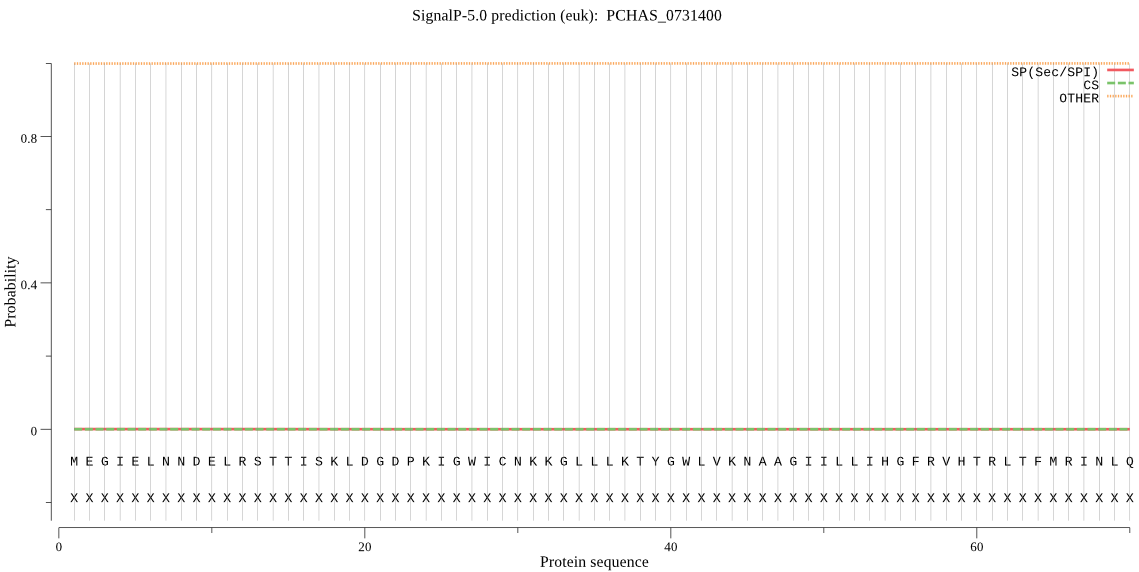
| PCHAS_0731400 | OTHER | 0.999812 | 0.000077 | 0.000112 |
MEGIELNNDELRSTTISKLDGDPKIGWICNKKGLLLKTYGWLVKNAAGIILLIHGFRVHT RLTFMRINLQMPNNNEDIVVDTNNYYIYKDSWIEKFNQNGYSVYALDLQGHGESQAWNNV RGEFSSFDDLVDDVIQYMNQIQDEISNDNQTNDDILNNNQTNDDILNNNQMNDESYDIVA TKKKKLPMYVIGYSMGGNIALRMLQVLNKEKGYNINTGESDNYKKCKAILDNSNHINEID NDMNNSKNDDPGTSSAGTSATANASTSDKHVEYDNYLDKFNIKGCISLSSMITLNETRNV GNILIKYLYLPVTSLLSRVAPNAKFPSDPDYKQFEYINNTYRYDKFRNPDETKFKYVYEL LKATVDVNYNINYMPKDIPLLFLHSSDDSVCHYQGTVSFHSKAKVETKNIYIFDKLDHAI TAAPGNEEVLNLIINWINNLRMNDGDKIKHEIEDKKKNEI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_0731400 | 17 S | STTISKLDG | 0.994 | unsp |