

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0800300 | OTHER | 0.999719 | 0.000060 | 0.000221 |
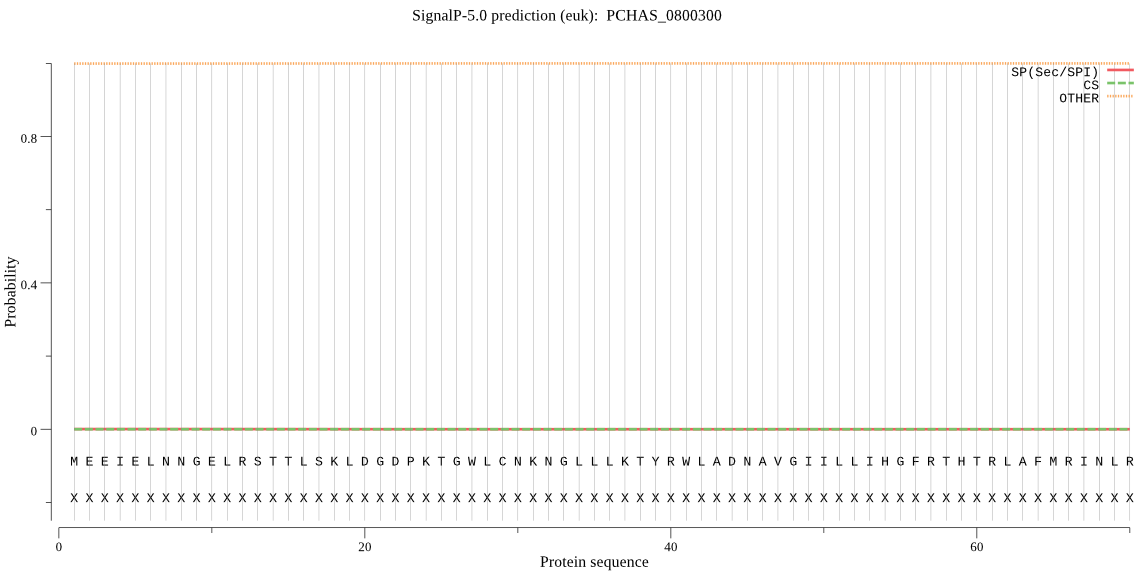
MEEIELNNGELRSTTLSKLDGDPKTGWLCNKNGLLLKTYRWLADNAVGIILLIHGFRTHT RLAFMRINLRMPNNNEGLIVDTNNYYIYKDSWIEKFNQNGYSVYALDLQGHGESQSQKNV KGDFSSYDDLVDDVLQYMNQIQDEISNDNQMNDESYDILTTKKKNLPMYVIGYSIGGNIA LRILQLLNKEKGNNINVGESDNNKKCNIMLDNSTNTNKIGKDMNNSNDYGSDNSCDSTSA TTNAIVNASDKHEGCNNCLDKFNIKGCITISGMIRIKSRLDPGNKSLKYFYLPLANFLSF VLPHVRIFSKSYKKSDYAANVSKYDKFRNHSGIKFKYIAEIIKATTALDYNINYMPKDIP LLFVHSKDDRVCSYEWTALFHNKVKVDKKDLHPVDGMGHATTIEPGNEDILKKIIDWIYD LRRNDEDEIEDG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0800300 | 278 S | IRIKSRLDP | 0.992 | unsp | PCHAS_0800300 | 278 S | IRIKSRLDP | 0.992 | unsp | PCHAS_0800300 | 278 S | IRIKSRLDP | 0.992 | unsp | PCHAS_0800300 | 311 S | IFSKSYKKS | 0.997 | unsp | PCHAS_0800300 | 373 S | DRVCSYEWT | 0.994 | unsp | PCHAS_0800300 | 17 S | STTLSKLDG | 0.995 | unsp | PCHAS_0800300 | 125 S | KGDFSSYDD | 0.991 | unsp |