

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_0814800 | SP | 0.379166 | 0.605811 | 0.015023 | CS pos: 21-22. IEG-FK. Pr: 0.9866 |
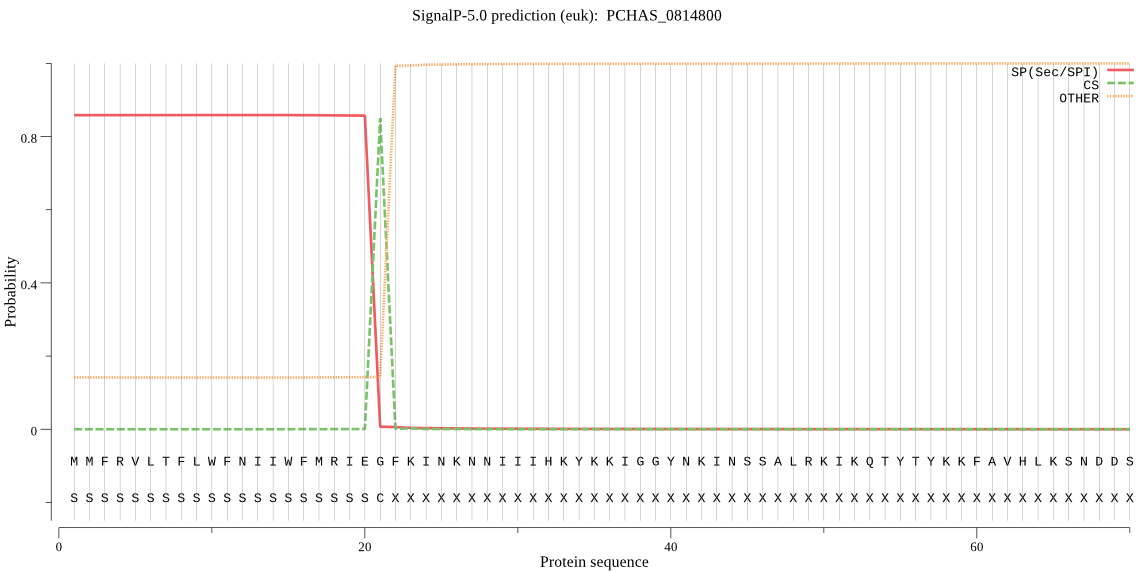
MMFRVLTFLWFNIIWFMRIEGFKINKNNIIIHKYKKIGGYNKINSSALRKIKQTYTYKKF AVHLKSNDDSVNNIKHRNIIKEQLSNFYKKNIYFFSKILLYMKNGASFITEKLTTSLASQ ILISISFFFLHFYLLSKNFIILLPYQLIPNHSNILMGLDFNNAFLIVSSLFFLKSFISNF ESFKTRLAPNRFTIPLKENKRKNILTVAVLFLASYILSGYVSIYAEQILRYSKSFNLDIS DTTIKSLQILIGHFMWVGCSIKIFSKFLYPYFKNNVSSLNFRYKDSWCFKVIYGYVCSHY IFNIIDLLNNTIINYFHNDDDIYIENSIDNIVNEKEFLSTFLCVISPCFSAPFFEEFIYR FFVLKSLSLFMNIHYAVTFSSLLFAIHHLNIFNVIPLFFLSFFWSYIYIYTDNILVTMII HSFWNIHVFLSSIYN