

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
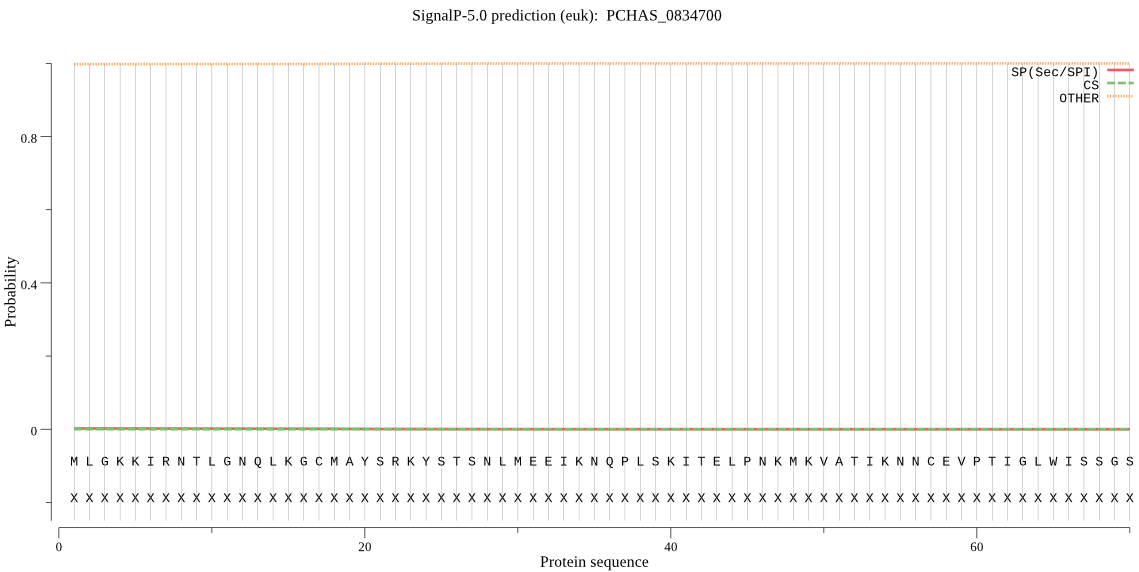
| PCHAS_0834700 | mTP | 0.141048 | 0.000011 | 0.858940 | CS pos: 24-25. RKY-ST. Pr: 0.8797 |
MLGKKIRNTLGNQLKGCMAYSRKYSTSNLMEEIKNQPLSKITELPNKMKVATIKNNCEVP TIGLWISSGSKYENKANNGVAHFLEHMIFKGTNKRNRVQLEKEIENMGAHLNAYTAREQT GYYFKCFKDDVKWCIELLSDILTNSVFDEKLIEMEKHVILREMEEVEKSADEVIFDKLHM TAFRDHPLGYTILGPVENIKNMKKNDILNYIQKNYTSDRMVLCAVGDVEHDNIVKLVEQN FSNIKPQDEKGLILKQEFDKIKPFFCGSEIIIRDDDSGPNAHVAVAFEGVPWTSSDSITF MLMQCIIGTYKKNEEGIVPGKLSANRTINNISNKMTIGCADYFTSFNTCYNNTGLFGFYV QCDELAVEHAVGELMFGITSLSYSITDEEVELAKIHLKTQLISMFESSSTLAEEISRQIL VYGRPITLAEFITRLNEIDAEEVKRVAWKYLHDRDIAVAAMGALHGMPQYFDLRQKTYWL RY
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_0834700 | 384 S | SLSYSITDE | 0.997 | unsp |