

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
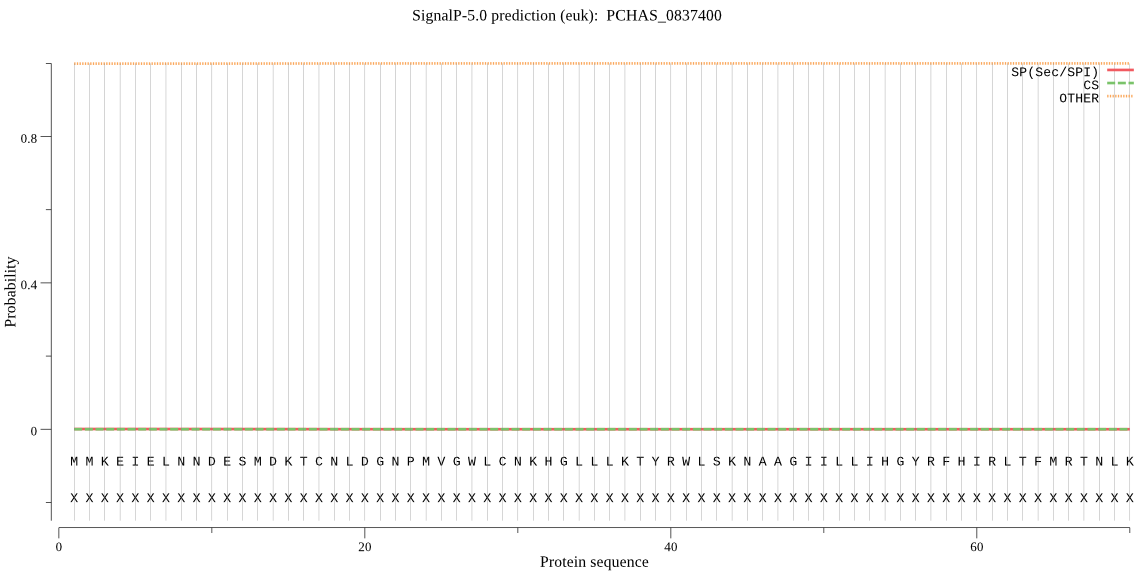
| PCHAS_0837400 | OTHER | 0.999917 | 0.000040 | 0.000043 |
MMKEIELNNDESMDKTCNLDGNPMVGWLCNKHGLLLKTYRWLSKNAAGIILLIHGYRFHI RLTFMRTNLKIPNHNESFVVDSNTYYIYKDSWIEKFNQSGYSVYGIDLQGHGESQSWKNV NGDFSRFDDLVDDVIQYMNQIHDEISNENQTDGESYDIVSTKKKRLPMYIIGYSMGANIA LRILQLLNKEKEDKIKTYNSNNYKKCSIMLNNSTDVNEIDNDMHNMNNSKYGDSGTSSGS TSATISAIVSTSDKHEGCYNDLDKFNIKGCASLSGMIRIKSIFNPGNISLKYFYLPIINF LSYILPNLQNLLEPHNNTSGYFSNICKYYIFRNINGKKYKYIYELAKATVTLDRNIKYMP KDVPLLFVHSQDDSICSYERAVAFYNKVNVSKKELYPVDGMDHNITTNPGNEEILKKVID WISDVRSNDEDGKENEL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_0837400 | 391 S | KVNVSKKEL | 0.994 | unsp |