

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
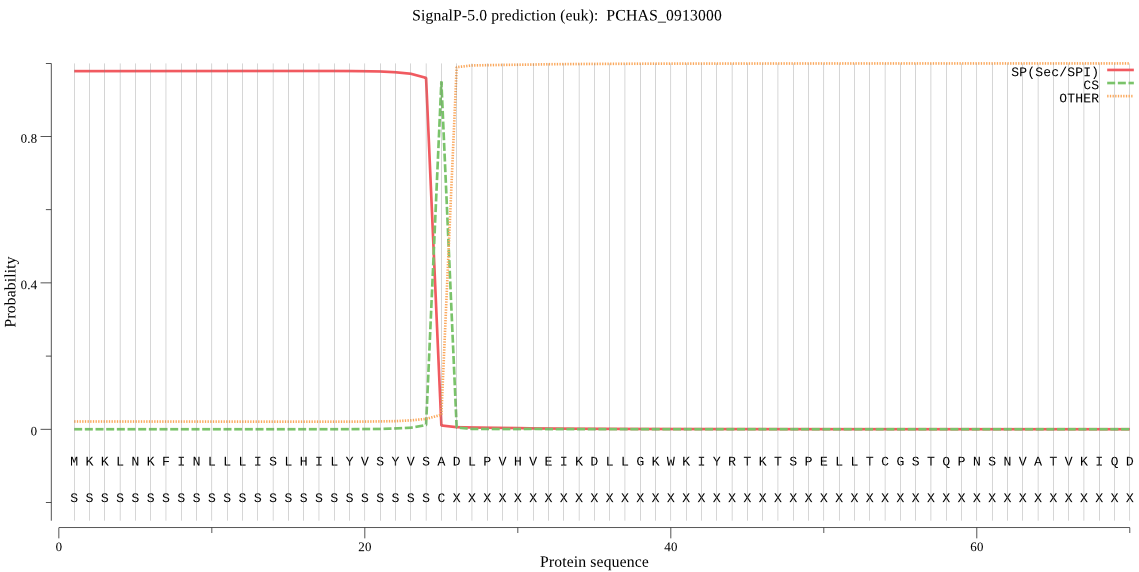
| PCHAS_0913000 | SP | 0.003002 | 0.996973 | 0.000025 | CS pos: 25-26. VSA-DL. Pr: 0.9708 |
MKKLNKFINLLLISLHILYVSYVSADLPVHVEIKDLLGKWKIYRTKTSPELLTCGSTQPN SNVATVKIQDYKKYLTDNHYIFSSELDVILSSDFVKYGEVHDITGNEHRKNWNVLAVYDE KKRKKIGTWTTIFDQGFEIRIANETYTGYMHYEPTGRCRAPSDEDITDSNGETVCYTTSH DKTRLGWVDIMYKNNEQSHGCFYAEKYDTVHEPNNYRSALNSSTSGRPEPVINDNKTPAS NQITNFDAYEPKTYIKADKVDYNKNSEMYWHKMKHSGNKKPLPEYMLKAQKQKYACPCNP NEDIDNERSDEDPDSPVSPNMIELGNSNVETNELDLNAYDEIKKAKHTELELNEMPKNFT WGDPFNDNVREYEVIDQLSCGSCYIASQMYVFKRRIEIGLTKLLDTKYNNDFDDALSLQT VLSCSFYDQGCHGGYPFLVSKMAKLQGIPLNSEFPYTARETKCPYPVNKGIPLSMMEADL IHKSESVTKDIKPSFREISGSPSIQNVAAEENSNENDLGDPNRWYAKEYNYVGGCYGCNQ CDGEKIIMNEIYRNGPVVGSIEVTPNFYEYVDGIYYDPGFPHAKKCTIDVHKDSSYVYNI TGWEKVNHAIVILGWGEQTIDGKLYKYWICRNSWGKTWGKEGYFKLIRGINYIAIENHAI YIDPDFTRGAGKILLDKMKKK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_0913000 | 162 S | CRAPSDEDI | 0.998 | unsp | PCHAS_0913000 | 309 S | DNERSDEDP | 0.998 | unsp |