

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
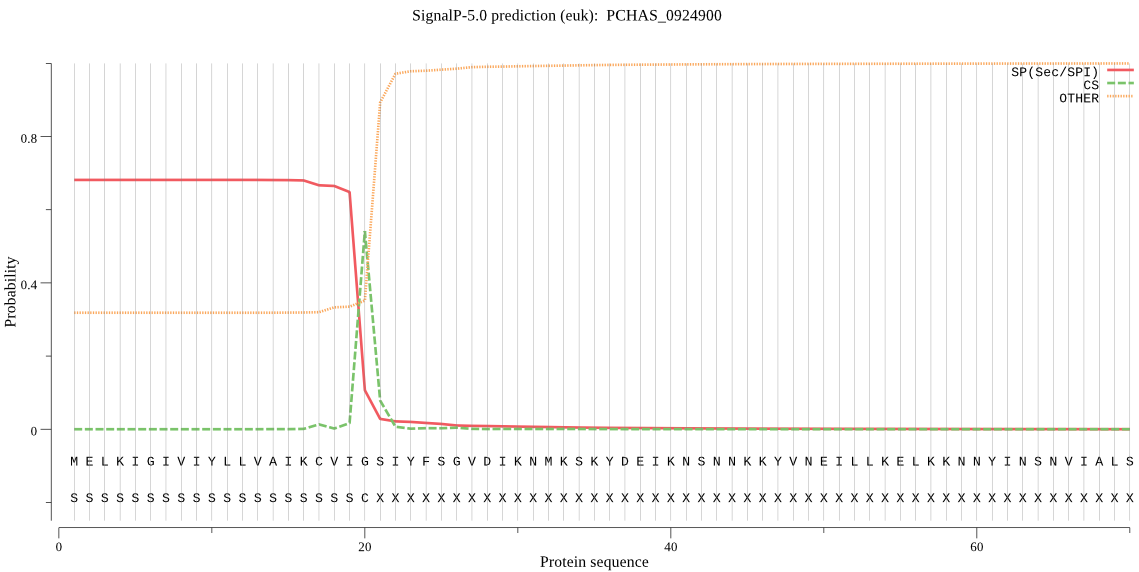
| PCHAS_0924900 | SP | 0.057176 | 0.942266 | 0.000557 | CS pos: 20-21. VIG-SI. Pr: 0.9407 |
MELKIGIVIYLLVAIKCVIGSIYFSGVDIKNMKSKYDEIKNSNNKKYVNEILLKELKKNN YINSNVIALSTSRHYFNYRHTANLLIAYKYLKNNGDIIDKNILLMVPFDQACNCRNIIEG TIFKKYEKFPNEYLNKNMEENLYNKLNIDYKNDNINDEQLRKIIRHRYNSFTPSKNRLYT NEYNEKNLFIYITGHGGINFLKIQEFNILSSSEFNLYIQELLIKNIYKNIFVVIDTCQGY SFYDDILAFINQNKIKNVFLLSSSDRNENSYSFFSSKYLSVSTVDRFTYNFFDYMENIHQ MQSKEIYKNLKLFSLQNILNYLKTKNLMSHPSINNSKFNVSSFLHDQNIIFYDSSMLFPK FFISKLNNTNDSNNNHDSVHSEHACFGYLGICGHIKSQIYLNMESLYHDRSYYNNDDVHN SYEIYFTDIGFNFKYFDIYYMITFLILLFFILLFLTLFVLN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_0924900 | 34 S | KNMKSKYDE | 0.995 | unsp |