

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
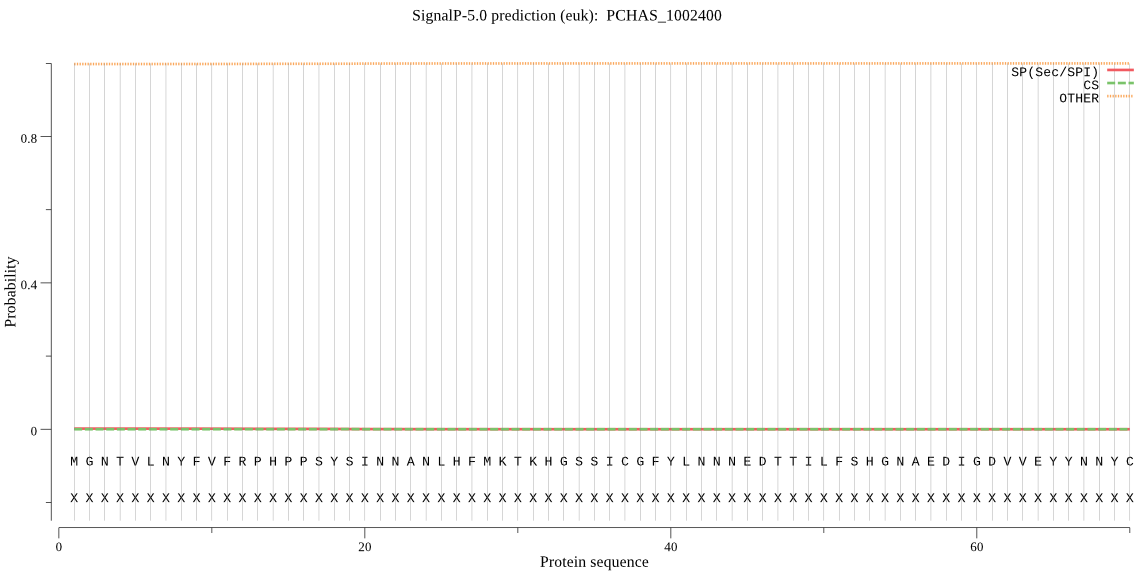
| PCHAS_1002400 | OTHER | 0.993368 | 0.002721 | 0.003911 |
MGNTVLNYFVFRPHPPSYSINNANLHFMKTKHGSSICGFYLNNNEDTTILFSHGNAEDIG DVVEYYNNYCKCIGVNMFLYDYSGYGHSTGYPSEEHVYNDVEAVYSYMTKTLCIPGGSIV AYGRSLGSTASVHIATKKKIKGLILQCPIASIHRVKLRLKSTLPFDFFCNIDKISNVKCP VLFIHGTNDTLIPYQGTVDMIMRTKVNTYYALIEGGGHNNLERCYYKQLHTSIFAFLHIL KTNVHEGMKISHDIASLSLFEFIKKYTLNEDPAQMKLKNVMNKIKKSNYETKPEKKRNSP LYRIDLNPPQLSDSENNDNIFKKSRESSNASRSYPSLTSISTIFDTDESTADGIPNKAFS ENSNNMSLEDISAFLCNNIPIRHLYKIRRWEKSIIENYNKNKKLEIDKNEIKELYRDYLY GPPDIDVYNFNNNNSYTCTKLENIKQNTKDSNFIGGSNTSKIGSSASKIGSNASKSGSNT SKTGSNASKSGSNTSKIGSNTSKTGSNASKTGSSASKSGSKIKSDNGKNKIIYDASGKLD GNNNSIASKKGEKNGSANYIYNQKKSMNTKSNLKSDNKQGLEKMPSNGASGSMSSTKREE LSSREGIANFCNIKSESNISSNMSATSDKKIIAKKGNKSRTNSHEKGGSNSSGNISVNDP PKSDASKCYWSDKVGNNKIEINKLKGKIQVNNNNISKYYETSPNETNFFDIPQENNALES SN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1002400 | 367 S | SNNMSLEDI | 0.991 | unsp | PCHAS_1002400 | 367 S | SNNMSLEDI | 0.991 | unsp | PCHAS_1002400 | 367 S | SNNMSLEDI | 0.991 | unsp | PCHAS_1002400 | 476 S | NASKSGSNT | 0.993 | unsp | PCHAS_1002400 | 490 S | NASKSGSNT | 0.993 | unsp | PCHAS_1002400 | 595 S | GSMSSTKRE | 0.995 | unsp | PCHAS_1002400 | 602 S | REELSSREG | 0.996 | unsp | PCHAS_1002400 | 643 S | SRTNSHEKG | 0.996 | unsp | PCHAS_1002400 | 656 S | SGNISVNDP | 0.991 | unsp | PCHAS_1002400 | 287 S | KIKKSNYET | 0.992 | unsp | PCHAS_1002400 | 327 S | KSRESSNAS | 0.996 | unsp |