

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
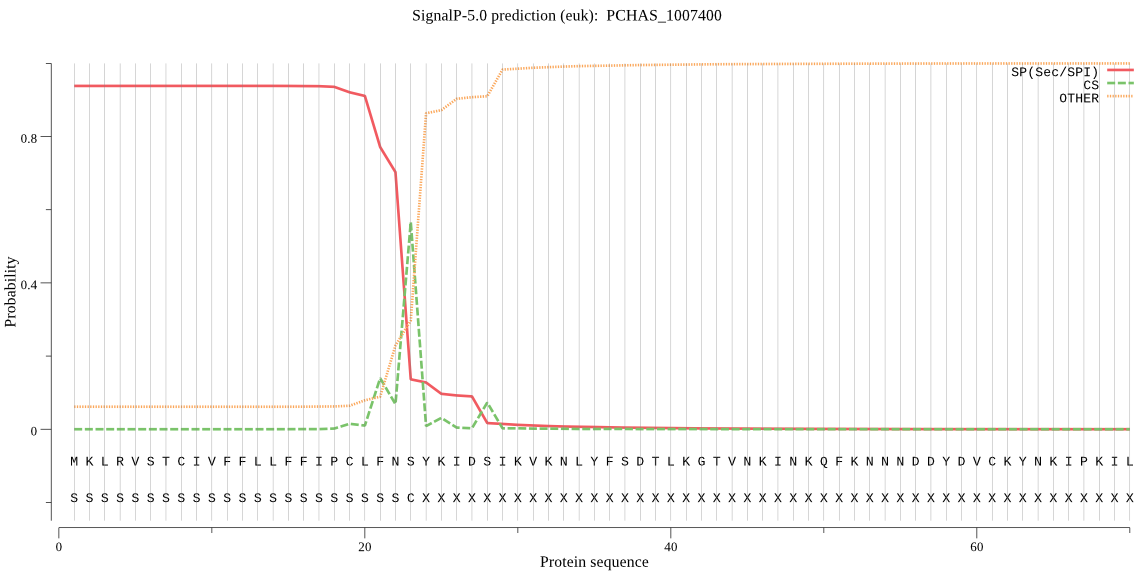
| PCHAS_1007400 | SP | 0.036795 | 0.963052 | 0.000154 | CS pos: 23-24. FNS-YK. Pr: 0.4841 |
MKLRVSTCIVFFLLFFIPCLFNSYKIDSIKVKNLYFSDTLKGTVNKINKQFKNNNDDYDV CKYNKIPKILIDRDKIKNNDKDSGYIVGIENTCDDTCVCIIDSKLRIIKHFIISHYKVVH KYGGVYPFFISSINSVFLNHYVNKILEGIDASKIKCFGFSVCPGISQNMDTARNYIGNFK KRHKHITASSVNHVYAHVLSPLFFSFYNDENTFTNNYEYKNESTKNDDPTDHINMNLFDT IKKDKIQMDKINSVLKVLNNNDVTKTKLKTITTEDFLNYGSYILKKKKIDDNNTKLDETK NDVNQVEGNNNMGKIPFSYIKNGYICVLVSGGSTQVYRVQKDKQNDINVCKISQTVDISI GDIIDKVARLLNLPVGLGGGPFLEQESEKYIKTLNGQKINEDVSFDLFEPFPVPFAPNNK INFSFSGILNHLSKIIKELKKDKNFENEKSKYAYYCQKYIFKHLLDQLNKIMYFSELHFN IKNIFIVGGVGCNKFLFESLKNLALNRNNIENQLKEYTKLKKRLKKKIKKIENSNFLTKK NPTPNDINDEIELRSSFAWNFYLKNLLKKKTANDILLALKAFDFDSFTKLKEKGSFLLQD QFLTSSVTKPWNVYRTPINLSRDNAAMICFNTLLNMHNKTNIHEDISEIKIKSTIKTQLQ NNFLLLSDIIVFDVFLDYFDHNLA
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1007400 | 223 S | YKNESTKND | 0.997 | unsp |