

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
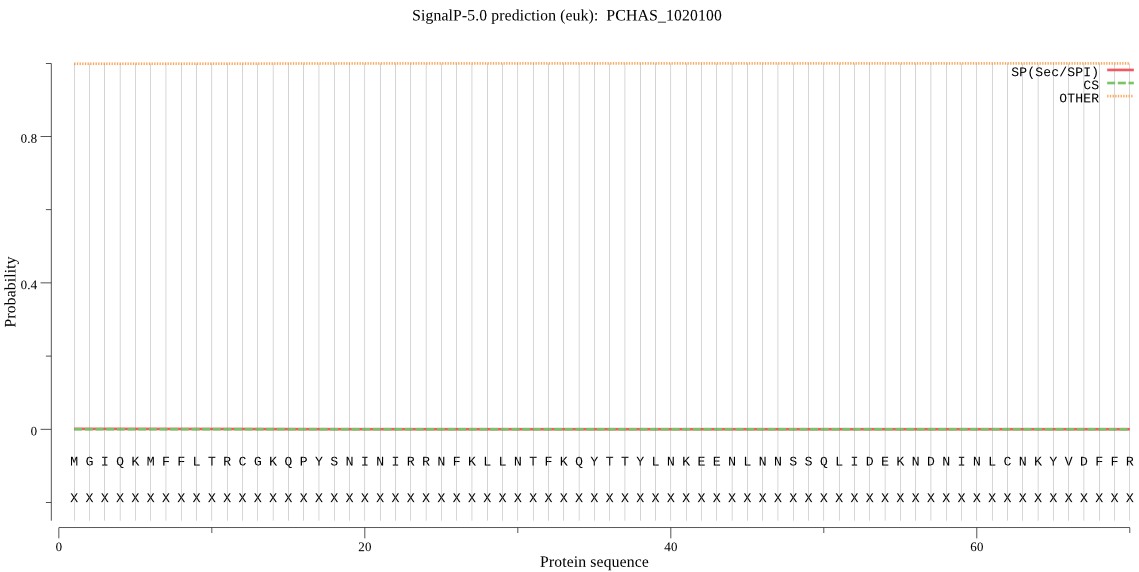
| PCHAS_1020100 | OTHER | 0.805939 | 0.000361 | 0.193700 |
MGIQKMFFLTRCGKQPYSNINIRRNFKLLNTFKQYTTYLNKEENLNNSSQLIDEKNDNIN LCNKYVDFFRDESRNIGIVTFKNICDKKNIFPNFLEELKNVIDHINNIISNEENNKFYIN EFKNKDNYLVKNLKKAIPYYDNKLKVLIINGSDTNFKKNNNTFLNSVDFNSYLKCDEGTN ADISSMFRIICNNIQQLPLITVSNINGICYNSGMDLILSTDFRISNENSKYGYDKTYIGL YPYGGSIQKLFRHIPMNYSKYLLLTSQTINAFDALKFNLIDVCLNQNEEFYINNSNVHFD HNLTKKNKIEIIKENIIKYFNDIFINENFKLKTNDDSFIFTLFFAFQFLFTPTYILQNIK LSINEGMLLSDSNAYLDCDRMVFEKSINTPERLEILNYLKTKVSEYKPKEN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1020100 | 225 S | DFRISNENS | 0.997 | unsp |