

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1033200 | OTHER | 0.999435 | 0.000140 | 0.000425 |
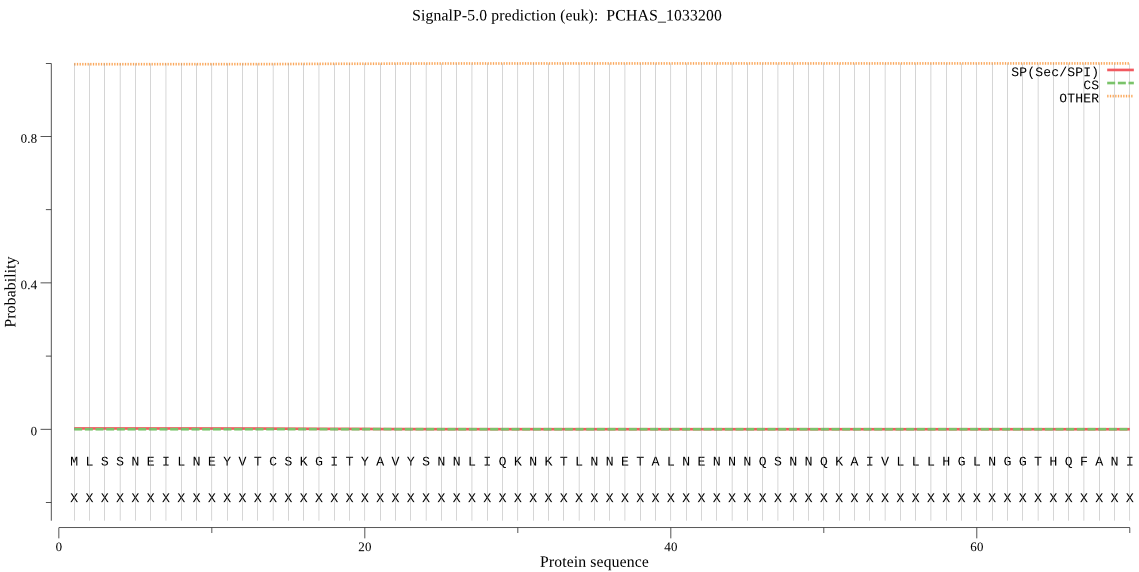
MLSSNEILNEYVTCSKGITYAVYSNNLIQKNKTLNNETALNENNNQSNNQKAIVLLLHGL NGGTHQFANIFKTLFDLDYQFISIDFYGHGNSSLFGNPNKYTEKLYTEQIYDVLKTKSLL NEKFIVIGFSMGCIIATHLSKDNKINIGKFCLISAAGMAKPRYRFLVFLLKYNIRLCLKL AKRYSRSVISEDTVKKEYYNFEKNFEEAAKQYEILKQNHEKFMETFLKVLIGIKLQNSKK YYFSLFKTNADVLFIYGKNDALTSYIYTQKFLEQKKEYSKNVKMIIIPECCHLVIHEKSH ELDHHLIYFLK