

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
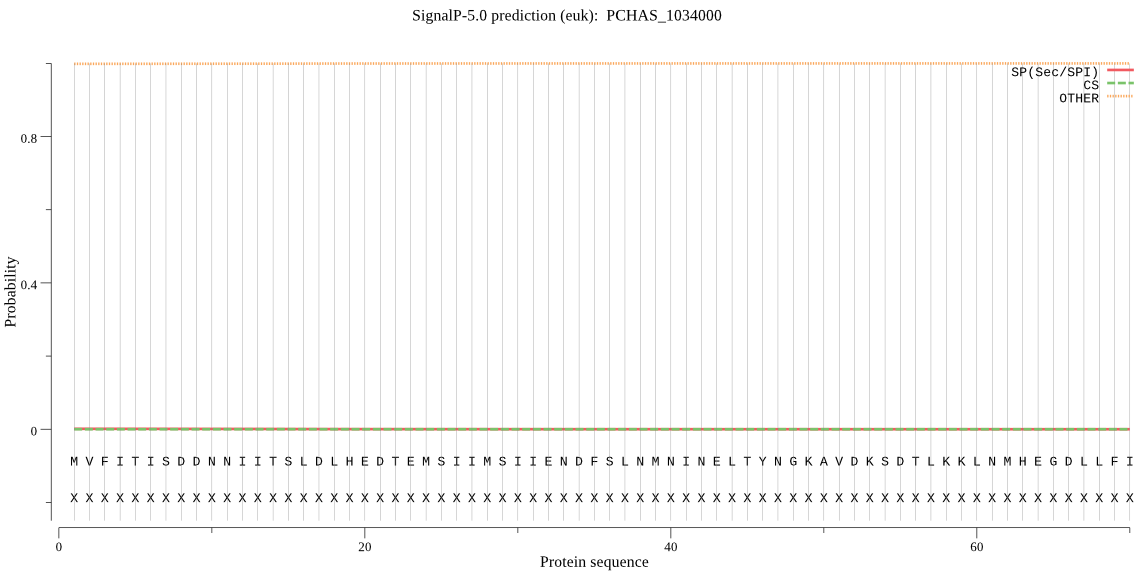
| PCHAS_1034000 | OTHER | 0.999986 | 0.000014 | 0.000001 |
MVFITISDDNNIITSLDLHEDTEMSIIMSIIENDFSLNMNINELTYNGKAVDKSDTLKKL NMHEGDLLFIRKKINLDMLQEELNANGFGNIMNNSGVTNTSATNSSTPNPGSILPSGLNN AGQGNNAAFNGILEQFRIFQEMEYIKKEAEKLLQLKTDKAKMSILQIQDKKLYDAINTAN LEEIKKIVKEKYEMEKKEKQREQEMYEKALKDPLSEESQKYIYEHIYKNQINSNLALAQE HFPEAFGVVYMLYIPVEINKNVIHAFVDSGAQTSIISKRCAEKCNILRLMDTRFTGIAKG VGTKSILGKIHMIDIKIGNYFYAVALTIIDDYDIDFIFGLDLLKRHQCSIDLKKNALVIE DNEIPFLAEKDIVKRSFESINLDSQ
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1034000 | 215 S | KDPLSEESQ | 0.995 | unsp |