

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1036400 | SP | 0.479501 | 0.483523 | 0.036976 | CS pos: 19-20. VLN-VR. Pr: 0.2759 |
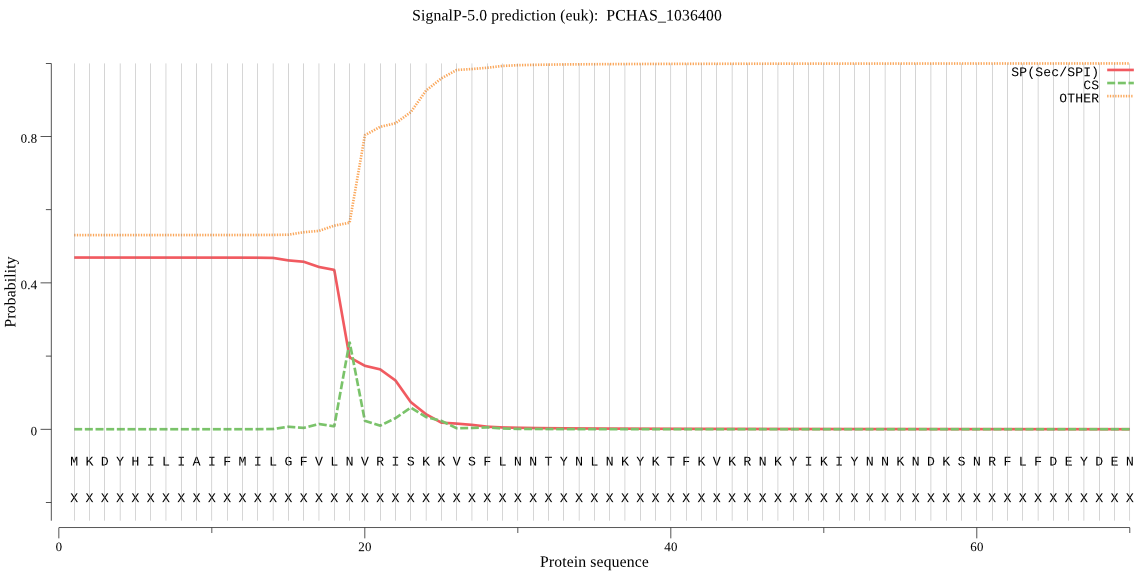
MKDYHILIAIFMILGFVLNVRISKKVSFLNNTYNLNKYKTFKVKRNKYIKIYNNKNDKSN RFLFDEYDENCIKALIMAREVAKNNNENEVLLKHLLVAILKTDSNIIEHIFKYFNISLHH FINKFNTEFDKNNLKKPEHESYDKNLAPSEQIDLQKSDTQNGDQENKLINNMNDKQIKDE EHNHIILNNLNNQSNDDITNRDDNTNADSTSSEHYGSEKVTDISNSYNNNADANYVANQN DDTFLVNNNIAKRNNQKVSKPNKEIKFSENCKLVLQNAILEARKKNKIFVNLTDILISMI NIAQENKECDFVKYLNELNISLSDLKKQINNYDENNLSSTLIHSPINKNIIPNDERHNNE HEIGNPNGDHYPDKQNRNMSDQPNNIINNMNNEYINQEKYFKNNEDNNFPQNNKFMPSVI TKDCLVDLVQRAYEKRMDNFFGRTDEIKRIIEILGRKKKSNPLLVGETGVGKTAIVEQLA YLILKNKVPYHLKNCKIFQLNLGNIVAGTKYRGEFEEKMKQILANISKDKKNILFIDEIH VIVGAGSGEGSLDASNILKPFLSSDNLQCIGTTTFQEYSKYIETDKALRRRFNCVNIKPL NSNETYKLLKKIKNSYEQYHNIYYTNEALKSIILLSDDYLPTLNFPDKAIDILDEAGSYQ KIKYERYITKKLRNENNIKTDPVKVEEKVDTNKADTINTASNQVENYSADVTSQVESNCL VDDLHMKYVTSDVIENIVSKKSSISFIKKNKKEEEKIQNLKEKLNKIIIGQEKVIDILSK YLFKAITNIKDANKPIGTLLLCGSSGVGKTLCAQVISQYLFNDDNIIVINMNEYIDKHSV SKLFGSYPGYVGYKEGGELTEAVKKKPFSIILFDEIEKAHSDVLHVLLQILDNGILTDSK GNKVSFKNTFIFMTTNVGSDIITDYFKVYNKNYANFGFKYYIKKEKNEKVEPQDTTNTNT HNGENKIQTDLDIFEEKIKTNKWYDELQPEIEEELKKKFLPEFLNRIDEKILFRQFLKRD IIQILENMIEDLKKRIKKRKKLNILIDDNIIKYICSDENNIYDINFGARSIRRALYKYIE DPIAAFLISNEYEPNDTMHLTLSPNNQINVNLLKHTITSLS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1036400 | 23 S | NVRISKKVS | 0.995 | unsp | PCHAS_1036400 | 217 S | EHYGSEKVT | 0.994 | unsp |