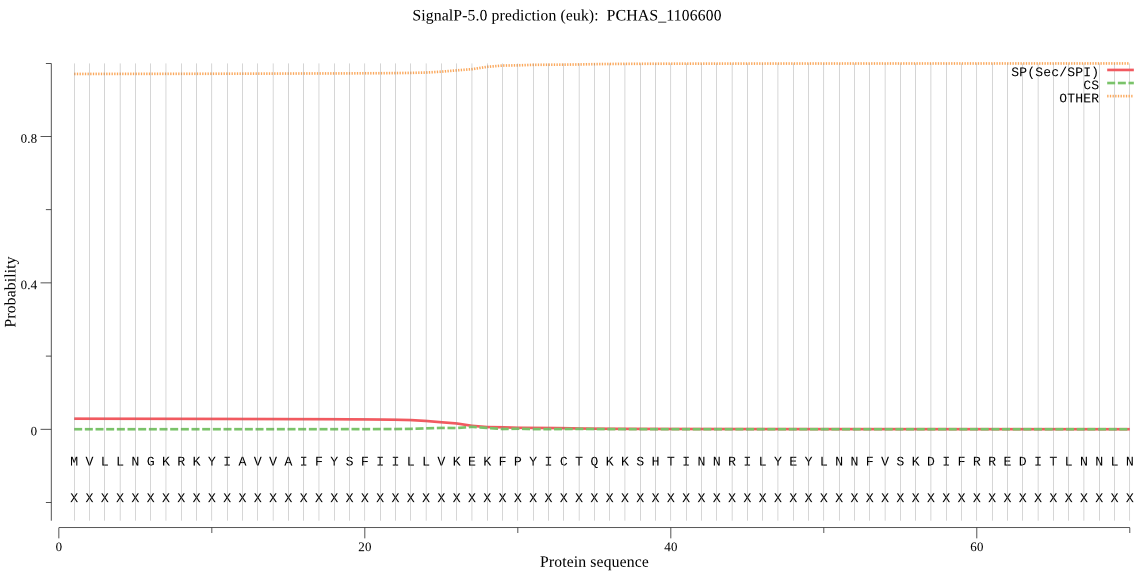
Fasta :-
MVLLNGKRKYIAVVAIFYSFIILLVKEKFPYICTQKKSHTINNRILYEYLNNFVSKDIFR
REDITLNNLNFVQTNLKYDKDVEIKENNDAQNADDNIFQRMYKFVLNFFYENKKDSTNKY
MKYGKYEHFNKINDIFEFMRNNGLPINITSVCLIDTGLDIKDEIINHFLNNDISTYNSDT
FHSVNINYKNADSFNFEINTENRDEDDYPEYESTFLENHNGDGSYEANSTSQGDPLKWGE
KKYKKGMDLQRSGIHVETCKAFDNSKESKNGSNIIPIIKCLEYCKTKNVKIIHIDYNINE
KNEELIEIIEDLKNSEIFLLLPSKGLLNEKSYEDNSVAYPSSFFENSENVFFIGSLNYLD
VFSYDVNIASNSQAQKNEYLKYRKDNVFLFDSINGSLKKGNDHDISYYEVRYSSAFFINI
LTTILNVYPNISINELRNMLSYSIIPEETDELKSQDIFEGNFEINKFIHGLLNGETNSSE
YVRKSKDVPSKEFTNKEFVSEDQEDDIVEAEVEVEAEAEVEAEAEVEAEVEAEAEVEAEV
EAEVEAEADSSADADYDKDGLDEDPINETDELDEPNEDSYEDNQSEKLLEDENYEFEEDP
SKDLHSVKENDIYVLENENPFNSSFMQISYDDKVRPKYVDNLDEGNYRPQYEMNRYPIIS
GDSLRGRQNMNNIQMVNDDIHYSENANNIEELYDNNFEGYRGNNLNQEYNKNYDNNKNVN
SEFGGLNNWRRNNEGSYLNDSEEVHHPFEWMDMQAGDIYRDKKNRLKENNYDNNETEIRR
RRKRGRGGENKRGRSMKNRNMRKTKKLGRDVKNRRYINRRTKRQIDKENKYNNSALKRQE
MKSYNNSQKAPKIIPRRYSR